Group leader: Enric Zelazny /Alexandre Martiniere
Key words: nanodomains, signaling, environment, membrane, abiotic, biotic, reactive oxygen species (ROS), Arabidopsis, cell biology, super resolution microscopy.
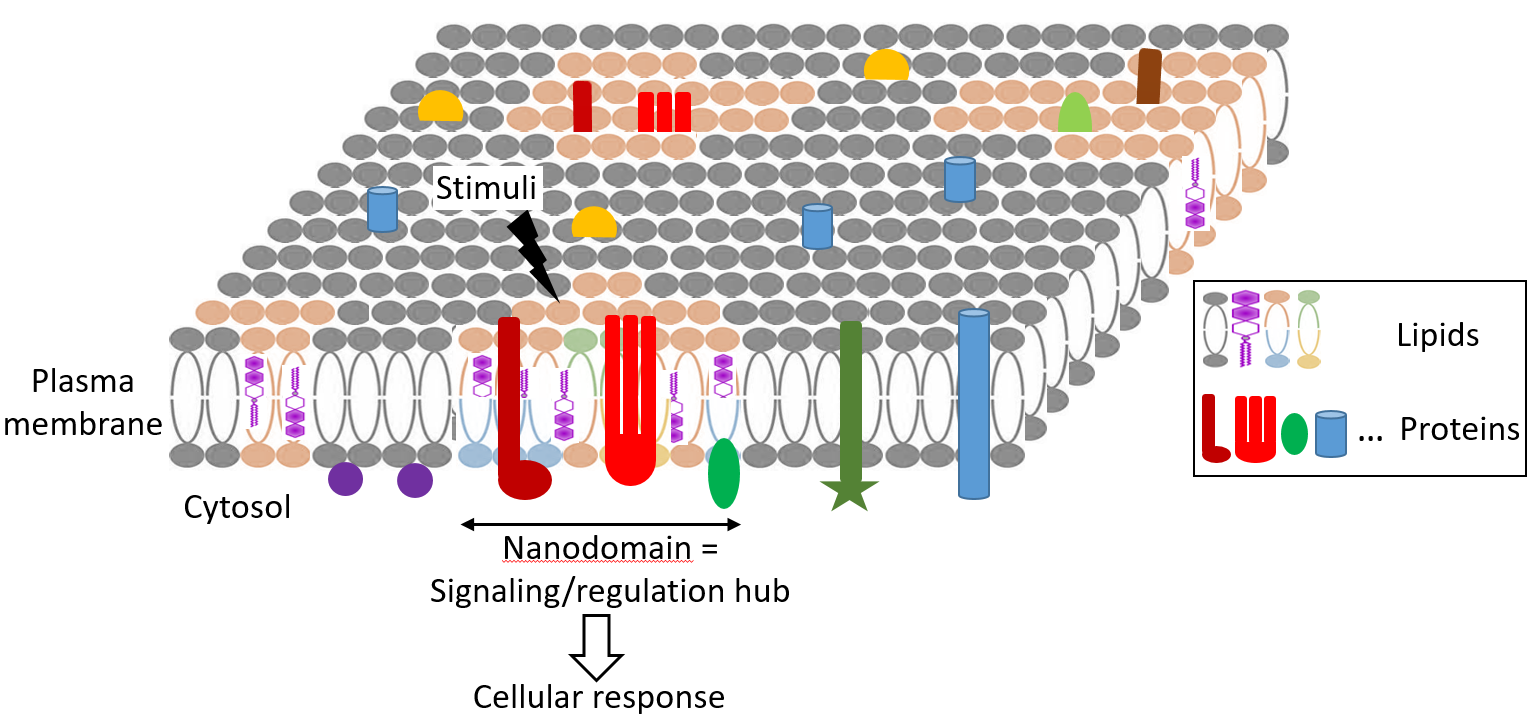
Presentation
Biological membranes contain a myriad of diverse domains with a nanometer scale size that correspond to the functional assemblies of specific proteins and lipids. The formation and maintenance of these nanodomains remain enigmatic, however it appears that these processes are dependent on the plasma membrane lipid composition, protein oligomerization and interactions with cytoskeletal and cell wall components. Although the function of nanodomains remains largely unknown, they emerge as signaling/regulation hubs that spatially and temporally coordinate critical cellular functions, notably through the establishment of protein-protein interactions. Thus, cell functioning relies on a ‘biochemical activity architecture’ through the specific arrangement of active molecules and their regulatory partners.
In this emerging scientific context, the objective of NANOPS is to study how membrane nanodomains can shape cell signaling processes, leading to appropriate plant responses to their environment, especially osmotic and immune stimuli, at various scales (cell, tissue, organism). To answer this question, we integrate interdisciplinary approaches including super resolution microscopy, biosensors imaging, transport assays, interactomic and lipidomic analyses, and modeling of protein and lipid organization.
Our current research axes are:
- Rho of Plant (ROP) nanodomains and establishment of cell signaling specificity during early plant osmotic signaling.
- Role of plasma membrane nanodomains containing Hypersensitive Induced Reaction (HIR) proteins in plant immunity.
- H2O2 nano environment formation and local decoding of ROS signaling.
Team members
Significant publications
Gorgues L, Smokvarska M, Mercier C, Igisch CP, Crabos A, Dongois A, Bayle V, Fiche J-B, Nacry P, Nollmann M, Jaillais Y, Martinière A✉ (2025) GEF14 acts as a specific activator of the plant osmotic signaling pathway by controlling ROP6 nanodomain formation. EMBO Rep., 26(8):2146-216
Hdedeh O*, Mercier C*, Poitout A*, Martinière A✉, Zelazny E✉ (2025) Membrane nanodomains to shape plant cellular functions and signaling. New Phytol., 245(4):1369-1385
Jaillais Y✉, Bayer EMF, Bergmann DC, Botella MA, Boutté Y, Bozkurt TO, Caillaud M-C, Germain V, Grossmann G, Heilmann I, Hemsley PA, Kirchhelle C, Martinière A, Miao Y, Mongrand S, Müller S, Noack LC, Oda Y, Ott T, Pan X, Pleskot R, Potocky M, Robert S, Sanchez Rodriguez C, Simon-Plas F, Russinova E, Van Damme D, Jaimie M. Van Norman, Weijers D, Yalovsky S, Yang Z, Zelazny E, Gronnier J✉ (2024) Guidelines for naming and studying plasma membrane domains in plants. Nat. Plants, 10(8):1172-1183
Smokvarska M, Bayle V, Maneta-Peyret L, Fouillen L, Poitout A, Dongois A, Fiche J-B, Gronnier J, Garcia J, Höfte H, Nolmann M, Zipfel C, Maurel C, Moreau P, Jaillais Y, Martinière A✉ (2023) The receptor kinase FERONIA regulates phosphatidylserine localization at the cell surface to modulate ROP signaling. Sci. Adv.![]() , 9(14):eadd4791
, 9(14):eadd4791
Martinière A✉, Zelazny E (2021) Membrane nanodomains and transport functions in plant. Plant Physiol., 187(4):1839-1855
Ugalde JM, Schlößer M, Dongois A, Martinière A, Meyer AJ✉ (2021) The latest HyPe(r) in plant H2O2 biosensing. Plant Physiol., 187(2):480-484
Bayle V*, Fiche J-B*, Burny C, Platre MP, Nollmann M, Martinière A✉, Jaillais Y✉ (2021) Single-particle tracking photoactivated localization microscopy of membrane proteins in living plant tissues. Nat. Protoc., 16(3):1600-1628
Martín Barranco A, Spielmann J, Dubeaux G, Vert G, Zelazny E✉ (2020) Dynamic control of the high-affinity iron uptake complex in root epidermal cells. Plant Physiol., 184(3):1236-1250
Smokvarska M*, Charbel F*, Platre MP*, Fiche J-B, Alcon C, Dumont X, Nacry P, Bayle V, Nollmann M, Maurel C, Jaillais Y, Martinière A✉ (2020) A plasma membrane nanodomain ensures signal specificity during osmotic signaling in plants. Curr. Biol., 30(23):4654-4664.e4
Platre MP, Bayle V, Armengot L, Bareille J, Marquès-Bueno MdM, Creff A, Maneta-Peyret L, Fiche J-B, Nollmann M, Miège C, Moreau P, Martinière A, Jaillais Y✉ (2019) Developmental control of plant Rho GTPase nano-organization by the lipid phosphatidylserine. Science, 364(6435):57-62
Martinière A*✉, Fiche J-B*, Smokvarska M, Mari S, Alcon C, Dumont X, Hematy K, Jaillais Y, Nollmann M, Maurel C (2019) Osmotic stress activates two reactive oxygen species pathways with distinct effects on protein nanodomains and diffusion. Plant Physiol., 179(4):1581-1593
Preprint
Danek M*, Hdedeh O*, Boutet J, Safi H, Abuzeineh A, Martín-Barranco A, Fiche J-B, Mercier C, Nollmann M, Boutté Y, Santoni V, Mongrand S, Martinière A, Zelazny E✉ (2025) Mechanisms controlling the plasma membrane targeting and the nanodomain organization of the plant SPFH protein HIR2. bioRxiv,
Collaborations
Lipid signaling
Marie-Cécile Caillaux (RDP-Lyon, France)
Yvon Jaillais (RDP-Lyon, France)
Yohann Boutté (LBM-Bordeaux, France)
Sébastien Mongrand (LBM-Bordeaux, France)
ROS biology
Andreas Meyer (Bonn University Germany)
Jean-Philippe Reichheld (LGDP, Perpignan, France)
Plant pathology
Laurent Noël (LIPME, Toulouse, France)
Corinne Audran (LIPME, Toulouse, France)
Thomas Kroj (PHIM, Montpellier, France)
Super resolution microscopy
Marcelo Nollmann (CBS, Montpellier, France)
Funding
ANR Nano-ROS (2025-2027, 559 k€)
ANR HIRAQIM (2024-2027, 547 k€)
I-Site HIROS (2024-2025, 15 k€)
ANR CellOsmo (2020-2024, 280 k€)
I-Site OptoSens (2023-2024, 99 k€)
ANR NUTRIR (2019-2023, 253 k€)
Former team members
Noah Didier, BTS, 2025
Omar Hdedeh, PhD, 2022-2024
Michal Danek, Post-doc, 2022-2023
Lucille Gorgues, PhD, 2021-2023
Héla Safi, ATER, 2021-2022
Armelle Dongois, Engineer, 2020-2021
Jessica Boutet, Engineer, 2021-2022
Anas Abuzeineh, Post-doc, 2020-2022
Marija Smokvarska, PhD, 2017-2020
