Genomics, Bioinformatics and Bio-analysis workshop

The Genomics, Bioinformatics and Bio-analysis (Gen2Bio) workshop is hosted by the IPSiM institute. Its aim is to provide support to UMR members with regard to data analysis (mainly sequence or expression analysis). This support can range from assistance with experimental design to the analysis of large ‘omic’ data sets.
The workshop helps with:
- Bioinformatics: assistance in choosing tools, developing analysis methods, and developing tools and databases.
- Bioanalysis: developing analysis pipelines and original custom scripts.
Whether for:
- Analysing RNA-seq, ChIP-seq or other omics data,
- Analysing and manipulating data (data mining, extracting relevant data from large datasets),
- Automating analyses.
Staff
Expertise
- RNA-seq: quality control, filters, mapping, counting, normalisation, differential expression analysis, visualisations, clustering, etc.
- ChIP-seq: quality control, mapping, peak detection, differential enrichment analysis, comparison with RNA-seq data
- Identification of mutations by DNA-seq
- Inference of regulatory networks and co-expression networks (random forests approaches -GENIE3/DIANE, co-expression, etc.)
- Data mining
- Sequence analysis: homology search, multiple alignments, phylogeny
- Pattern search (cis sequences, restriction sites, etc.), search for polymorphic markers, synteny
- Search for protein domains, protein-protein interactions, subcellular localisation, etc.
Equipment / Technologies
We mainly use scripting languages (R, PHP, Perl, SQL). We have a Debian server at our disposal within the Laboratory and, if necessary, we use the GenoToul or ISDM computing servers.
Online tools developed by IPSiM members:
- DIANE, a Shiny application for RNA-seq data analysis developed by Océane Cassan, a doctoral student in the Sirene team: https://diane.ipsim.inrae.fr/
- AraNoisy, a Shiny application for exploring the level of inter-individual transcriptional variability for Arabidopsis thaliana genes developed by Sandra Cortijo, researcher in the Sirene team: https://jlgroup.shinyapps.io/AraNoisy/
Training
Since 2023, the Gen2Bio workshop has been leading the IPSiM data analysis discussion group, which meets every two months. The purpose of this group is to discuss and learn about various topics related to data analysis. The topics covered include (non-exhaustive list): the use of AI in coding learning, data standardisation, the choice of statistical tests, R and Rmarkdown, best practices in graph creation, and RNA-seq data analysis.
Sandra Cortijo has also created the following two training courses (in French), which are available to all UMR staff:
- Introduction to data analysis with R: https://scortijo.github.io/2024_L3_R/
- RNA-seq data analysis with R: https://scortijo.github.io/2024_M2_RNAseq/
Publications
Mozzanino T, Li M, Fizames C, Adamo M, Lejay L, Dubos C, Platre M, Martin A✉ (2025) An intracellular transporter mitigates the CO2-induced decline in iron content in Arabidopsis shoots. Febs Lett., (in press)
Lecuyer C*, Vettor A*, Fizames C, Javot H, Martin A, Mazouzi M, Cortijo S✉ (2025) Establishment, maintenance and consequences of inter-individual transcriptional variability for a gene involved in nitrate nutrition in plants. bioRxiv,
Hmidi D, Muraya F, Fizames C, Véry A-A, Roelfsema MR✉ (2025) Potassium extrusion by plant cells: evolution from an emergency valve to a driver of long-distance transport. New Phytol., 245(1):69-87
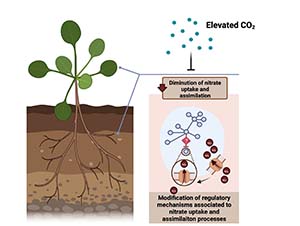
Cassan O, Pimparé L-L, Mozzanino T, Fizames C, Devidal S, Roux F, Milcu A, Lèbre S, Gojon A, Martin A✉ (2024) Natural genetic variation underlying the negative effect of elevated CO2 on ionome composition in Arabidopsis thaliana. eLife, 23(12):RP90170
Corratgé-Faillie C, Matic L, Chmaiss L, Zhour H, Lolivier J-P, Audebert P-A, Bui TX, Seck M, Chinachanta K, Fizames C, Wipf D, Sentenac H, Véry A-A, Courty P-E, Luu DT✉ (2023) Arbuscular mycorrhizal fungus Rhizophagus irregularis expresses an outwardly Shaker-like channel involved in rice potassium nutrition. bioRxiv,
Chaput V, Li J, Séré D, Tillard P, Fizames C, Moyano TC, Zuo K, Martin A, Gutiérrez RA, Gojon A, Lejay L✉ (2023) Characterization of the signalling pathways involved in the repression of root nitrate uptake by nitrate in Arabidopsis thaliana. J. Exp. Bot., 74(14):4244-4258
Zhour H, Bray F*, Dandache I*, Marti G, Flament S, Perez A, Lis M, Cabrera-Bosquet L, Perez T, Fizames C, Baudoin E, Madani I, El Zein L, Véry A-A, Rolando C, Sentenac H, Chokr A, Peltier J-B✉ (2022) Wild wheat rhizosphere-associated plant growth-promoting bacteria exudates: effect on root development in modern wheat and composition. Int. J. Mol. Sci., 23(23):15248
Séré D, Cassan O, Bellegarde F, Fizames C, Boucherez J, Schivre G, Azevedo J, Lagrange T, Gojon A, Martin A✉ (2022) Loss of Polycomb proteins CLF and LHP1 leads to excessive RNA degradation in Arabidopsis. J. Exp. Bot., 73(16):5400-5413
Yang S, Nguyen TH, Fizames C, Li J, Wang S, Vernet A, Guiderdoni E, Wang S, Huang Y-N, Hao D-L, Wang J, Sentenac H, Véry A-A, Su Y-H✉ (2022) A short shaker channel mediates K+ loading to root conducting tissues and contributes to rice grain yield under field conditions. Res. Square,
Ruffel S*, Chaput V*, Przybyla-Toscano J, Fayos I, Ibarra C, Moyano TC, Fizames C, Tillard P, O’Brien JA, Gutiérrez RA, Gojon A, Lejay L✉ (2021) Genome-wide analysis in response to nitrogen and carbon identifies regulators for root AtNRT2 transporters. Plant Physiol., 186(1):696-714
Garcia K✉, Guerrero-Galán C, Frank HER, Haider MZ, Delteil A, Conéjéro G, Lambilliotte R, Fizames C, Sentenac H, Zimmermann SD✉ (2020) Fungal Shaker-like channels beyond cellular K+ homeostasis: A role in ectomycorrhizal symbiosis between Hebeloma cylindrosporum and Pinus pinaster. PLoS one, 15(11): e0242739
Jacquot A, Chaput V, Mauriès A, Li Z, Tillard P, Fizames C, Bonillo P, Bellegarde F, Laugier E, Santoni V, Hem S, Martin A, Gojon A, Schulze W, Lejay L✉ (2020) NRT2.1 C-terminus phosphorylation prevents root high affinity nitrate uptake activity in Arabidopsis thaliana. New Phytol., 228(3):1038-1054