The first record of the parasitism of Pyemotes sp. (Acari: Pyemotidae) on Eurytoma schreineri Schreiner, 1908 (Hymenoptera: Eurytomidae) inside plum seeds
Tonka, Tomáš  1
; Pultar, Oldřich
1
; Pultar, Oldřich  2
and Ouředníčková, Jana
2
and Ouředníčková, Jana  3
3
1✉ University of South Bohemia in České Budějovice, Faculty of Agriculture and Technology, Department of Plant Production, Studentská 1668, 370 05 České Budějovice, Czech Republic.
2Research and Breeding Institute of Pomology Holovousy Ltd., Holovousy 129, 508 01 Holovousy, Czech Republic.
3Research and Breeding Institute of Pomology Holovousy Ltd., Holovousy 129, 508 01 Holovousy, Czech Republic.
2025 - Volume: 65 Issue: 4 pages: 1011-1017
https://doi.org/10.24349/al9k-nfqlOriginal research
Keywords
Abstract
Introduction
Mites of the family Pyemotidae (Oudemans, 1937) are described as parasites of a large number of insects including members of the order Hymenoptera (Macías-Macías and Otero-Colina, 2004), Lepidoptera (da Cunha et al. 2006; Oliveira et al. 2007), and Coleoptera (de Sousa et al. 2020; da Silva et al. 2022). Two ecological groups based on the behavior towards the host have been recognized: the scolyti group and the ventricosus group, containing the most important species, Pyemotes tritici (LaGrèze-Fossat & Montagné, 1851), P. ventricosus (Newport, 1850) and P. herfsi (Oudemans, 1936), considered conspecific with P. zwoelferi (Krczal 1963) (Cross and Moser 1975; Cross et al. 1981). Pyemotid mites are also well known as straw-itch mites, causing dermatitis in humans (Stingeni et al. 2023), however, despite medical concerns, mites have been evaluated for decades as possible biological control agent for their potential use against insect and arthropod pests (Yu et al. 2010; Chen et al. 2021; Hervet and Morrison 2021). The mass production of these mites and their biocontrol potential have been shown to be effective in laboratory assays; however, the efficacy of mass releases for pest control under field conditions remains uncertain (Bruce and LeCato 1979; Thorvilson et al. 1987; Hoschele and Tanigoshi 1993). However, it has been demonstrated that pyemotid mites are able to destroy some insect populations successfully (Li et al. 2019; Tian et al. 2020).
Pyemotid mites are traditionally placed within the infraorder Eleutherengona in the order Trombidiformes (Ye et al. 2022). The phylogenetic position of family Pyemotidae in mite lineages is mainly based on the morphological or behavioral traits, such as evolution of parasitism or feeding habits (Kaliszewski et al. 1995). Nevertheless, the taxonomic position of pyemotid mites and their phylogenetic relationships with closely related mite groups remain unclear due to the lack of well-supported molecular data, which is essential for resolving the phylogeny of Pyemotes spp.
Plum seed wasp (PSW), Eurytoma schreineri (Hymenoptera: Eurytomidae), is an important polyphagous pest which can cause considerable damage to the stone fruits in the Palearctic region. The larvae of the wasp feed inside the plum seed, causing massive fall and drying of the infected fruits.
Here we report Pyemotes mite, that was determined based on the molecular analysis of the 18S rDNA gene, parasitizing the plum seed wasp, E. schreineri, in the Czech Republic. To our knowledge, this is the first description of a pyemotid species infecting plum seed wasp larvae and pupae within the plum endocarp. Our study reveals a parasitic relationship between pyemotid mites and the immature stages of the plum seed wasp. However, further research is necessary to confirm the potential of these mites for controlling plum seed wasps, both in the laboratory and under natural field conditions.
Material and methods
The initial study was focused on the evaluation of PSW pathogens using a molecular approach, in conjunction with research on plant pathogens that may be transmitted by E. schreineri. In early March, when diapause begun to terminate, mummified plums were collected from the ground below trees of the Prunus domestica L. variety in an orchard in the vicinity of Osvětimany, Czech Republic. The collected samples were transferred to the laboratory, where the infested fruits were maintained at outdoor temperatures until the end of April to simulate natural conditions for larval development. The pits were then shattered, and the larvae from the cracked pits were kept individually in a dark chamber to await pupation and the emergence of the adult wasp. They were checked for infection daily and physogastric females were collected from the surface of infested plum seed wasp larvae and pupae and preserved in 70% ethanol for further analysis.
To confirm the mite species, we used molecular methods. DNA was extracted from individual specimens using the DNeasy Blood and Tissue kit (Quiagen) following the manufacturer's procedure, with final elution in 50 µl of buffer AE. To amplify the target mitochondrial cytochrome c oxidase subunit I (COI) gene region, the widely used universal primers LCO1490 (5′-GGTCAACAAATCATAAAGATATTGG-3′) and HCO2198 (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′) were selected (Folmer et al., 1994). In addition, the forward primer eukss18f (5′-CACCAGGTTGATTCTGCC-3′) and the reverse primer 1492R (5′-GGGTTACCTTGTTACGACTT-3′) were used to amplify the small subunit 18S rRNA gene region (https://www.mrdnalab.com/16s-ribosomal-sequencing.html ![]() ). PCR was performed in 20 µl total volume containing 4 µl of DNA, 1 µl of each primer, 10 µl of goTaq MasterMix (Promega), and 4 µl nuclease-free water. For each PCR reaction, a negative control containing sterile distilled water was included. The amplification was performed in a Biometra TAdvanced cycler (Biometra, Germany) by the initial denaturation at 94 °C for 3 min followed by 35 cycles of denaturation at 94 °C for 45 s, annealing at 45 °C for 30 s, elongation at 72 °C for 90 s and a final elongation period at 72 °C for 10 minutes, using the eukss18f/1492r primers. The PCR conditions for the COI gene amplification using the LCO1490/HCO2198 primers were as described in the original study by Folmer et al. (1994).
). PCR was performed in 20 µl total volume containing 4 µl of DNA, 1 µl of each primer, 10 µl of goTaq MasterMix (Promega), and 4 µl nuclease-free water. For each PCR reaction, a negative control containing sterile distilled water was included. The amplification was performed in a Biometra TAdvanced cycler (Biometra, Germany) by the initial denaturation at 94 °C for 3 min followed by 35 cycles of denaturation at 94 °C for 45 s, annealing at 45 °C for 30 s, elongation at 72 °C for 90 s and a final elongation period at 72 °C for 10 minutes, using the eukss18f/1492r primers. The PCR conditions for the COI gene amplification using the LCO1490/HCO2198 primers were as described in the original study by Folmer et al. (1994).
Positive PCR products were sequenced at SEQme s.r.o. (Czech Republic). The obtained sequences were then compared against the BLAST database to identify them to the closest taxonomic match based on sequence similarity. For genus-level identification of mites, we used the highest-scoring 18S rRNA gene matches. The species-level determination requires supporting analyses using additional genetic markers (e.g., COI, ITS2) or morphological data due to the conserved nature of 18S rRNA gene region.
Results and discussion
From 526 collected PSW larvae and pupae, 127 (24.14%) were parasitized by pyemotid mites. Among the infected individuals, 113 (21.48%) and 14 (2.66%) were in the larval and pupal phases, respectively (Figure 1A, B). The mites were found on the cuticle and intersegmental sites of the host, visible as yellow-white physogastric females with spherical opisthosoma. Specimen identification was based on molecular methods, specifically PCR amplification of extracted DNA, followed by sequencing and BLAST search. We extracted DNA from twenty specimens, specifically physogastric females. Despite multiple attempts, we were unable to amplify the COI region using the LCO1490/HCO2198 primers. However, we successfully amplified 18S rRNA region from two samples, which were subsequently sequenced and deposited in GenBank. The results indicated that the amplified gene sequences from pyemotid mites infecting PSW (accession numbers: PQ349768, PV367228) shared ≥ 99% identity (with 100% query coverage in both cases) with Pyemotes herfsi 18S ribosomal RNA gene sequence (KY922162). Amplification of the COI gene region with the primers developed by Folmer et al. (1994) failed because the effectiveness of these primers is sometimes low based on our experience. This also corresponds to the small number of Pyemotes spp. COI sequences deposited in the GenBank database.
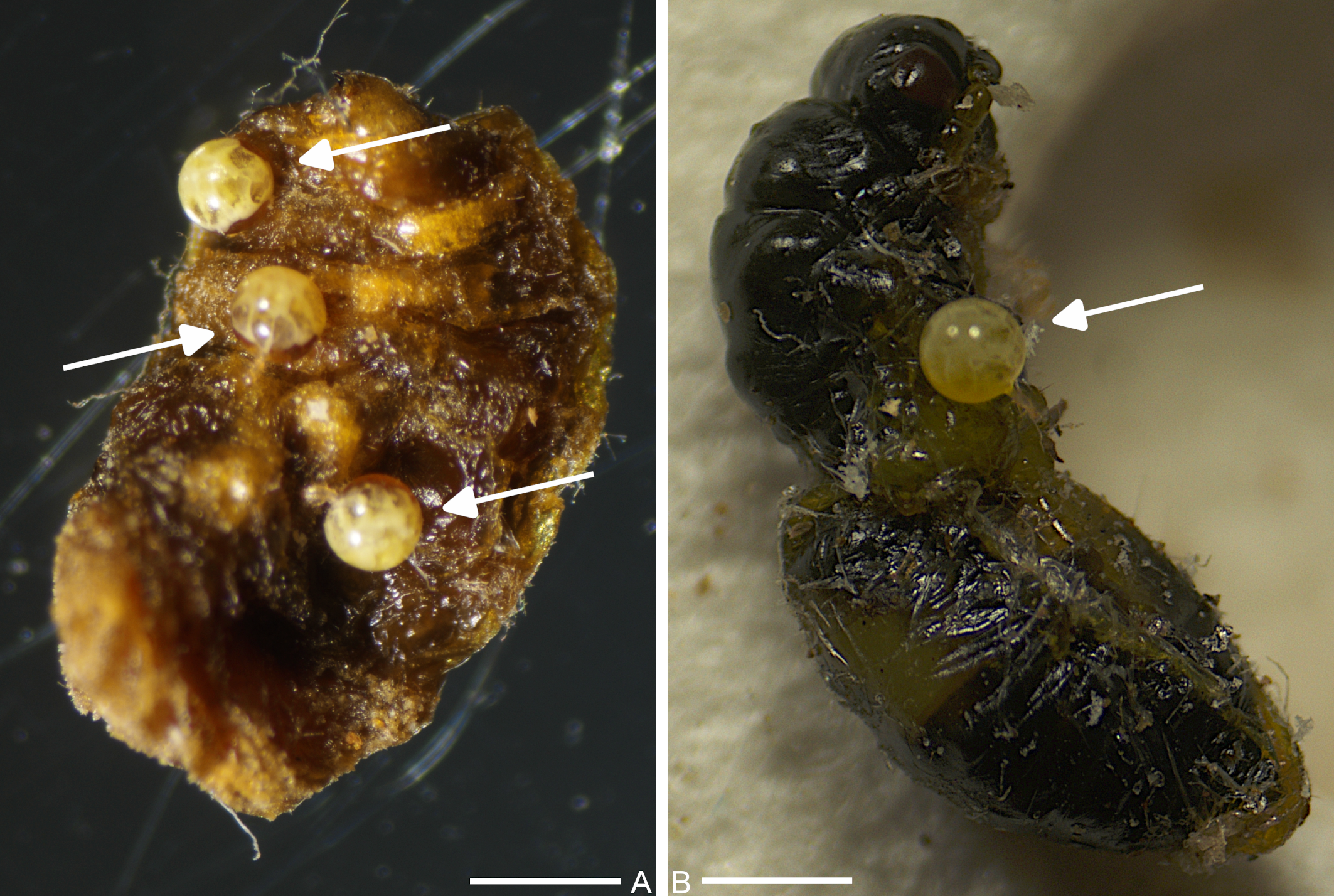


The mites that parasitized E. schreineri larvae belong to the ventricosus group, which is associated with a wide range of insect hosts. Among them, P. tritici, ventricosus and herfsi are the most common species, usually producing toxins that induce immobilization of insect prey (Weiser and Sláma 1964; Tomalski et al. 1988). To the best of our knowledge, there are no previous records of Pyemotes spp. infecting insects in the Czech Republic, either in the field or in the laboratory, although P. zwoelferi (considered a synonym of P. herfsi), originally collected in France, has been reared for laboratory assays (Weiser 1963). Additionally, pyemotid mites have been occasionally found in stored grains (Hubert et al. 2006), and a case of mass dermatitis caused by P. herfsi has been described in a food mixing shed (Samšiňák et al. 1979). Therefore, the results described in this study represents both the first report of pyemotid mites parasitism on E. schreineri larvae and pupae, and the first reported natural infection by Pyemotes sp. on insect hosts in the Czech Republic.
Although pyemotid mites are serious parasites of humans and natural enemies of important agricultural pests, DNA molecular analyses are rare for this group, and only one complete mitochondrial genome of P. zhonghuajia was sequenced (Ye et al. 2022). With lack of molecular data, the phylogenetic relationships among higher-order mite lineages remain essentially unresolved, with no clear indication of what the closest relatives of pyemotid mites are. Genetic studies could also highlight possible cryptic species diversity within the traditionally recognized, even well-known, genus Pyemotes. In pyemotid mites, species identification is routinely based on morphological character sets (Krczal 1959). There are many species within the genus Pyemotes that are morphologically very similar and therefore difficult to distinguish (Cross et al. 1981). Further research requires increased efforts integrating molecular and morphological data to obtain additional information that would support the classification of the family Pyemotidae and the position of the genus Pyemotes in the mite phylogenetic lineages (Pepato and Klimov 2015).
Our molecular analysis showed that 18S rRNA gene sequences isolated from PSW in the Czech Republic have a high similarity with the P. herfsi sequence deposited in the GenBank database (KY922162), a strong indicator that we may deal with the same species. However, the limited molecular data from GenBank (only three sequences deposited only) avoid to make definitive conclusions about pyemotid species described here. We therefore provisionally classify this mite at the genus level (Pyemotes sp.). Although molecular data suggest a high probability to be P. herfsi, conclusive species assignment requires additional morphological and molecular analyses to eliminate all taxonomic uncertainty.
As with 28S, the ability to distinguish closely related taxa using regions of 18S ribosomal genes is generally limited (e.g. Tang et al. 2012). Since both genes exhibit slower evolutionary rates, congeneric taxa may posses very similar, if not identical, copies of these genes (Hillis and Dixon 1991). Closely related taxa are most reliably distinguished using gene regions with slightly faster evolutionary rates. Therefore, the mitochondrial COI gene is widely used in taxonomic studies due to its relatively high evolutionary rate, which helps differentiate closely allied species (Muller 2006; Machida and Tsuda 2010). However, we were unable to successfully amplify the COI gene from the extracted DNA. We acknowledge that species determination based on the 18S rRNA gene has limitations, as does sequence similarity analysis relying only on the single 18S sequence of P. herfsi deposited in GenBank (KY922162). Given the lack of additional data, species-level conclusions based on the sequence similarity of 18S gene analyses must remain tentative.
Little is known about the KY922162 sequence (obtained from P. herfsi collected in the USA), except its origin. We assume that its taxonomic identification was based on morphological examination of slide-mounted mite specimen (Klimov et al. 2018). Therefore, if the P. herfsi 18S reference sequence (KY922162) was obtained from specimen that was morphologically identified as P. herfsi, we have no doubt to believe that our sequences (PQ349768, PV367228) also belong to P. herfsi - despite our lack of expertise in mite morphology and taxonomic identification. However, it is widely accepted that the 18S rRNA gene is slow evolving and typically not suitable for species-level taxonomy, thus we tentatively classify our sequences as Pyemotes sp. Further analyses using faster evolving markers (e.g., COI, ITS), supplemented with morphological data from other specimens, are required to confirm species identity, and resolve potential cryptic diversity within Pyemotes groups.
One remaining concern involves potential taxonomic ambiguity. In North America, P. tritici has been reported parasitizing several coleopteran hosts, including Agrilus auroguttatus and A. coxalis, closely related species of A. planipennis (Coleman et al. 2015). It is assumed that P. herfsi is a European species that was introduced to North America (Broce et al. 2006). Most non-European records of P. herfsi are linked to human dermatitis (Polter et al. 2025), while records of insect infections generally lack associated host data (but see Broce et al. 2006; Klimov et al. 2018 for exceptions). Interestingly, since the erection of the P. ventricosus species group, there have been no reports of P. tritici, the most commonly reported species of the ventricosus group, infecting insects in Europe.
If P. herfsi is believed to be a European species, its reported occurrence in North America has been confirmed through both morphological identification and the validity of its 18S sequence deposited in GenBank. However, only three Pyemotes 18S rRNA sequences (now including our two records) are currently available in database. Given this limited dataset, robust Pyemotes species discrimination remains challenging, highlighting the critical need for additional research utilizing expanded genetic sampling, and rapidly evolving genetic markers, such as COI or ITS, to resolve taxonomic ambiguities within Pyemotes groups.
We isolated DNA from twenty individual specimens. It is evident that the DNA concentration obtained from a single physogastric female is very low. Therefore, we succeeded in amplifying and sequencing only two samples, both of which were identified as small subunit ribosomal RNA gene sequences, now deposited in GenBank. These results suggest that PCR success depends more on primer binding efficiency rather than the initial DNA quantity. This implies that even with limited amount of DNA, effective primer design is critical for amplification. To ensure sequence reliability, we deliberately avoided DNA extraction from bulk samples (containing multiple individuals) due to concerns about the accuracy of the resulting sequences. While bulk samples might provide higher DNA concentrations, intraspecific variability among individual specimens could affect the clarity and quality of the obtained sequences.
In conclusion, there remains a significant gap in molecular analyses and phylogenetic studies of pyemotid mites. Future research is needed to obtain Pyemotes spp. genomic data using next-generation sequencing (NGS) or whole-genome sequencing (WGS). These approaches could bring new data on the phylogenetic relationships within the family Pyemotidae, and the evolutionary origin and taxonomic position of pyemotid mites in the acariform phylogeny in general. Unlike PCR amplification which relies on known, conserved primers, genomic data obtained through NGS technology may provide novel phylogenetic markers or more phylogenetically informative sites. Furthermore, genome datasets provide additional information to population genetic analyses and functional gene investigations.
Despite the fact that there are some concerns that pyemotid mites can have a potentially serious impact on human health, multiple studies recommend ways to incorporate these mites into the control of agricultural and forest pests. A short life cycle, a wide range of hosts, and rapid population growth with high reproductive potential are hallmarks of highly efficient Pyemotes spp. used in biological control (He et al. 2019). It is likely that pyemotid mites are present in PSW populations in nature; however, further research needs to be done to use Pyemotes spp. to control seed wasps. Beyond that, the questions regarding which environmental factors affect mite population and what the mite threshold level is for efficient control of seed wasps remain unanswered. A fundamental understanding of mite-host interactions underpins the development of better control strategies.
Acknowledgments
This research was financially supported by the Ministry of Agriculture of the Czech Republic (QK22020019).
References
- Broce A. B., Zurek L., Kalisch J. A., Brown R., Keith D. L., Gordon D., Goedeke J., Welbourn C., Moser J., Ochoa R., Azziz-Baumgartner E., Yip F., Weber J. 2006. Pyemotes herfsi (Acari: Pyemotidae), a mite new to North America as the cause of bite outbreaks. J. Med. Entomol., 43: 610-613. https://doi.org/10.1093/jmedent/43.3.610
- Bruce W. A., LeCato G. L. 1979. Pyemotes tritici: potential biological control agent of stored product insects. In Rodriguez J. (Ed.). Recent Advances in Acarology. Academic Press. p. 213-220. https://doi.org/10.1016/B978-0-12-592201-2.50032-4
- Chen Y.-C., Tian T.-A., Chen Y.-H., Yu L.-C., Hu J.-F., Yu X.-F., Liu J.-F., Yang M.-F. 2021. The biocontrol agent Pyemotes zhonghuajia has the highest lethal weight ratio compared with its prey and the most dramatic body weight change during pregnancy. Insects: 12:490. https://doi.org/10.3390/insects12060490
- Coleman T., Jones M., Hoddle M., Haavik L., Moser J., Flint M., Seybold S. 2015. Pyemotes tritici (Acari: Pyemotidae): A parasitoid of Agrilus auroguttatus and Agrilus coxalis (Coleoptera: Buprestidae) in the southwestern United States of America and southern Mexico. Canad. Entomol., 147: 1-5. https://doi.org/10.4039/tce.2014.38
- Cross E. A., Moser J. C. 1975. A new, dimorphic species of Pyemotes and a key to previously described forms (Acarina: Tarsonemoidea). Ann. Entomol. Soc. America, 68: 723-732. https://doi.org/10.1093/aesa/68.4.723
- Cross E. A., Moser J. C., Rack G. 1981. Some new forms of Pyemotes (Acarina: Pyemotidae) from forest insects, with remarks on polymorphism. Int. J. Acarol., 7: 179-196. https://doi.org/10.1080/01647958108683260
- da Cunha U. S., Silva E. S. de Moraes, G. J., Vendramim J. D. 2006. Occurrence of the mite Pyemotes sp. (Acari: Pyemotidae) in insect rearing in laboratory. Neotrop. Entomol., 35: 563-565. https://doi.org/10.1590/S1519-566X2006000400023
- da Silva C. A. D., de Moraes G. J., Castilho R. C., Ramalho F. S., Lima T. A. 2022. New parasitism record of Pyemotes tritici (LaGreze-Fossat & Montagne, 1851) (Acari: Pyemotidae) on boll weevils inside cotton squares. Acarologia, 62: 426-430. https://doi.org/10.24349/lldq-iy5f
- de Sousa A. H., Mendonça G. R. Q., Lopes L. M., Faroni L. R. D. 2020. Widespread infestation of Pyemotes tritici (Acari: Pyemotidae) in colonies of seven species of stored-product insects. Genet. Mol. Res., 19: gmr18548. https://doi.org/10.4238/gmr18548
- Folmer O., Black M., Hoeh W., Lutz R., Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Marine Biol. Biotechnol., 3(5): 294-299.
- He L., Li L., Yu L., He X. Z., Jiao R., Xu C., Zhang L., Liu J. 2019. Optimizing cold storage of the ectoparasitic mite Pyemotes zhonghuajia (Acari: Pyemotidae), an efficient biological control agent of stem borers. Exp. App. Acarol., 78: 327-342. https://doi.org/10.1007/s10493-019-00386-0
- Hervet V. A. D., Morrison W. R. 2021. Prospects for use of biological control of insect and mites for the food industry in North America. Agronomy, 11: 1969. https://doi.org/10.3390/agronomy11101969
- Hillis D. M., Dixon M. T. 1991. Ribosomal DNA: molecular evolution and phylogenetic inference. Q. Rev. Biol., 66: 411-453. https://doi.org/10.1086/417338
- Hoschele W., Tanigoshi L. K. 1993. Pyemotes tritici (Acari: Pyemotidae), a potential biological control agent of Anagasta kuehniella (Lepidoptera: Pyralidae). Exp. App. Acarol., 17: 781-792. https://doi.org/10.1007/BF00225851
- Hubert J., Münzbergová Z., Kučerová Z., Stejskal V. 2006. Comparison of communities of stored product mites in grain mass and grain residues in the Czech Republic. Exp. Appl. Acarol., 39: 149-158. https://doi.org/10.1007/s10493-006-0026-y
- Kaliszewski M., Athias-Binche F., Lindquist E. E. 1995. Parasitism and parasitoidism in Tarsonemina (Acari: Heterostigmata) and evolutionary considerations. In Baker J. R., Muller R., Rollinson D. (Eds.). Adv. Parasitol. Academic Press, 35: 335-367. https://doi.org/10.1016/S0065-308X(08)60074-3
- Klimov P. B., OConnor B. M., Chetverikov P. E., Bolton S. J., Pepato A. R., Mortazavi A. L., Tolstikov A. V., Bauchan G. R., Ochoa R. 2018. Comprehensive phylogeny of acariform mites (Acariformes) provides insights on the origin of the four-legged mites (Eriophyoidea), a long branch. Mol. Phylogenet. Evol., 119: 105-117. https://doi.org/10.1016/j.ympev.2017.10.017
- Krczal H. 1959. Systematik und Ökologie der Pyemotiden. In Stammer, H. J. (Ed.). Beiträge Zur Systematik Und Ökologie Mitteleuropäischer Acarina, Vol 2. Leipzig: Akademische Verlagsgesellschaft Geest & Portig K.-G. p. 385-465.
- Li L., He L., Yu L., He X. Z., Xu C., Jiao R., Zhang L., Liu J. 2019. Preliminary study on the potential of Pyemotes zhonghuajia (Acari: Pyemotidae) in biological control of Aphis citricola (Hemiptera: Aphididae). Syst. Appl. Acarol., 24: 1116-1120. https://doi.org/10.11158/saa.24.6.12
- Machida R. J., Tsuda A. 2010. Dissimilarity of species and forms of planktonic Neocalanus copepods using mitochondrial COI, 12S, nuclear ITS, and 28S gene sequences. PLoS One, 28: e10278. https://doi.org/10.1371/journal.pone.0010278
- Macías-Macías J., Otero-Colina G. 2004. Infestation of Pyemotes tritici (Acari: Pyemotidae) on Melipona colimana (Hymenoptera: Apidae: Meliponinae): A case study. Agrociencia, 38: 525-528.
- Mueller R. L. 2006. Evolutionary rates, divergence dates, and the performance of mitochondrial genes in Bayesian phylogenetic analysis. Syst. Biol., 55: 289-300. https://doi.org/10.1080/10635150500541672
- Oliveira C., Matos C., Hatano E. 2007. Occurrence of Pyemotes sp. on Tuta absoluta (Meyrick). Braz. Arch. Biol. Technol., 50. https://doi.org/10.1590/S1516-89132007000700003
- Pepato A.R., Klimov P.B. 2015. Origin and higher-level diversification of acariform mites - evidence from nuclear ribosomal genes, extensive taxon sampling, and secondary structure alignment. BMC Evol. Biol., 15: 178. https://doi.org/10.1186/s12862-015-0458-2
- Polter E. J., McCoy K., Lee X., Bedno S. A. 2025. Dermatitis outbreak associated with Pyemotes herfsi mites among animal shelter workers and volunteers in Wisconsin, a case study from September 2023. Int. J. Environ. Health Res., 1-8. https://doi.org/10.1080/09603123.2025.2515534
- Samšiňák K., Chmela J., Vobrázková E. 1979. Pyemotes herfsi (Oudemans, 1936) as causative agent of another mass dermatitis in Europe (Acari, Pyemotidae). Folia Parasitol., 26: 51-54.
- Stingeni L., Hansel K., Casciola G., Bianchi L., Tramontana M., et al. 2023. Human ectoparasitosis by mites of the genus Pyemotes Amerling 1861 (Acarina: Pyemotidae). Italian J. Dermatol. Venereol., 158: 4-14. https://doi.org/10.23736/S2784-8671.22.07481-3
- Tang C Q., Leasi F., Obertegger U., Kieneke A., Barraclough T. G., Fontaneto D. 2012. The widely used small subunit 18S rDNA molecule greatly underestimates true diversity in biodiversity surveys of the meiofauna, Proc. Natl. Acad. Sci. U.S.A., 109: 16208-16212. https://doi.org/10.1073/pnas.1209160109
- Thorvilson H. G., Phillips, Jr. S. A., Sorensen A. A., Trostle M. R. 1987. The straw itch mite, Pyemotes tritici (Acari: Pyemotidae), as a biological control agent of red imported fire ants, Solenopsis invicta (Hymenoptera: Formicidae). Florida Entomol., 70: 439-444. https://doi.org/10.2307/3494785
- Tian T., Yu L., Sun G., Yu X., Li L., Wu C., Chen Y., Yang M., Liu J. 2020. Biological control efficiency of an ectoparasitic mite Pyemotes zhonghuajia on oriental armyworm Mythimna separata. Syst. Appl. Acarol., 25: 1683-1692. https://doi.org/10.11158/saa.25.9.13
- Tomalski M. D., Bruce W. A., Travis J., Blum M. S. 1988. Preliminary characterization of toxins from the straw itch mite, Pyemotes tritici, which induce paralysis in the larvae of a moth. Toxicon, 26: 127-132. https://doi.org/10.1016/0041-0101(88)90164-X
- Weiser J. 1963. Über Massenzuchten von Pyemotes-Milben (Acarina). Beitr. zur Entomol., 13: 547-551. https://doi.org/10.21248/contrib.entomol.13.3-4.547-551
- Weiser J., Sláma K. 1964. Effects of the toxin of Pyemotes (Acarina: Pyemotidae) on the insect prey, with special reference to respiration. Ann. Entomol. Soc. America, 57: 479-482. https://doi.org/10.1093/aesa/57.4.479
- Ye S., Zhang H.-Y., Song Y.-F., Yang M.-F., Li L.-T., Yu L.-C., Liu J.-F. 2022. Complete mitochondrial genome of Pyemotes zhonghuajia (Acari: Pyemotidae). Sys. Appl. Acarol., 27: 1677-1686. https://doi.org/10.11158/saa.27.8.16
- Yu L., Zhang Z.-Q., He L. 2010. Two new species of Pyemotes closely related to P. tritici (Acari: Pyemotidae). Zootaxa, 2723: 1-40. https://doi.org/10.11646/zootaxa.2723.1.1



2024-09-26
Date accepted:
2025-09-23
Date published:
2025-10-03
Edited by:
Roy, Lise

This work is licensed under a Creative Commons Attribution 4.0 International License
2025 Tonka, Tomáš; Pultar, Oldřich and Ouředníčková, Jana
Download the citation
RIS with abstract
(Zotero, Endnote, Reference Manager, ProCite, RefWorks, Mendeley)
RIS without abstract
BIB
(Zotero, BibTeX)
TXT
(PubMed, Txt)



