First cytogenetic data on two trombidioid mites (Prostigmata) from Türkiye, with a checklist of known chromosome numbers in mites
Karağaç, Rümeysa  1
; Eroğlu, Halil Erhan
1
; Eroğlu, Halil Erhan  2
; Hekimoğlu, Olcay
2
; Hekimoğlu, Olcay  3
and Sevsay, Sevgi
3
and Sevsay, Sevgi  4
4
1Department of Biology, Institute of Science, Erzincan Binali Yıldırım University, Erzincan, Türkiye.
2Department of Biology, Faculty of Sciences and Arts, Yozgat Bozok University, Yozgat, Türkiye.
3Department of Biology, Faculty of Science, Division of Ecology, Hacettepe University, Ankara, Türkiye.
4✉ Department of Biology, Faculty of Sciences and Arts, Erzincan Binali Yıldırım University, Erzincan, Türkiye.
2025 - Volume: 65 Issue: 2 pages: 589-601
https://doi.org/10.24349/1srq-1vejOriginal research
Keywords
Abstract
Introduction
Studies on determining the chromosome numbers of mites began many years ago. Although the first chromosome studies on mites began with Reuter's (1909) work on the oogenesis of Pediculopsis graminum, the most detailed cytological study was Schrader's (1923) research on Tetranychus bimaculatus from the Tetranychidae family (Reuter 1909; Schrader 1923). These studies continued with tarsonemid, gamasid and phytoseiid mites (Cooper 1937, 1939; Hansell et al. 1964; Oliver 1964, 1965, 1972, 1973, 1977; Patau 1934; Sokolov 1934, 1945 1954; Warren 1940; Wysoki 1973; Wysoki and Swirski 1968). Subsequent studies on Tetranychus urticae aimed to gain a deeper understanding of the biology and reproductive characteristics of these harmful mite species to develop biological control strategies (Helle and Bolland 1967). Later, chromosome studies were extended to ticks (Ixodoidea) (Oliver et al. 1973) and other mite groups (Bolland and Ueckermann 1984; Eroğlu and Per 2016; Gümüş et al. 2022; Heethoff et al. 2006; Onrat et al. 2006; Wysoki and Bolland 1983; Zalewska 1979). Specific chromosomal studies on eriophyoid mites emerged later, making them one of the first mite groups to undergo detailed cytogenetic analysis (Norton et al. 1993).
Although detailed chromosome number data are lacking for most species within the Trombidiidae family, some studies have focused on the biological characteristics of the group at the genus level. Members of this family are known to be taxonomically complex, with morphological similarities that complicate classification. Notably, species in this group exhibit a distinct life cycle: they are characterized by a complex life cycle which includes heteromorphic parasitic larvae and predatory post-larval instars (Wohltmann and Wendt 1996). However, in contrast to the available biological and life cycle data, chromosomal studies in Trombidiidae are exceedingly rare. To date, chromosomal data have been reported only for Allothrombium fuliginosum, with a diploid chromosome number of 2n=24 in both males and females (Sokolov 1954).
In general, Trombidium species exhibit high morphological similarity, which makes species-level identification challenging without the preparation of detailed microscope slides. In such morphologically cryptic groups, molecular identification—particularly DNA barcoding using the COI gene—has proven to be an effective complementary tool for distinguishing closely related species (Blaxter 2004; Davrat 2005; Knowlton 2000). Therefore, the integration of both morphological and molecular data is crucial for accurate species delimitation in Trombidiidae. The aim of this study is to provide the first cytogenetic characterization of two trombidioid mite species, Trombidium latum and Eutrombidium trigonum, and to support the identification of T. latum using mitochondrial COI barcoding data. We analyzed the chromosome numbers of T. latum from the Trombidiidae family in both the larval and adult stages, and the post-larval stage of E. trigonum from the Microtrombidiidae family. Additionally, a comprehensive list of mite species with known chromosome numbers to date is provided, serving as a valuable reference for future taxonomic and cytogenetic studies.
Materials and methods
Trombidioid mites sampling and locations
Samples were collected from Erzincan, Türkiye and preserved and prepared according to the methods described by Mąkol (2005) and Mąkol and Sevsay (2011). Specimens of E. trigonum were collected from Ahmediye village, 39°52′54″N, 39°20′31″E, 2048 m, moist soil and moss, 27 April 2021. Specimens of T. latum were collected from Demirpınar Stream Basin, Üzümlü Town. 39°57′12″N, 39°35′21″E, from soil close to the Astragalus, 22 May 2021 and from the Ahmediye village, the sampling site of E. trigonum.
Larvae of T. latum were obtained by rearing a field-collected ovigerous female under controlled laboratory conditions, solely for the purpose of obtaining specimens for cytogenetic analysis. The female was placed in a glass vial containing charcoaled Plaster-of-Paris and covered with a tight, semi-transparent lid to maintain humidity and allow visual inspection. No experimental data were recorded from this procedure beyond larval availability. The morphological identification follows Mąkol (2002) for T. latum, and Gabryś (1999) for E. trigonum.
Cytogenetic method
The cytogenetic preparations were made from the protocol presented by Imai et al. (1988) and modified by Gokhman and Quicke (1995). The samples were pretreated and crushed in hypotonic solution (1% sodium citrate containing 0.005% colchicine) for 30 min., and transferred into a fresh hypotonic solution (0.075 M KCl) for 20 min. Then the samples were fixed in freshly prepared fixative solution (acetic acid: ethanol - 3:1). After the fixation, the material was transferred to a clean slide and air-dried. Then the slides were stained with 8% Giemsa.
Ten metaphase plates with good distribution and prominent chromosomes were evaluated for each species. The plates were imaged using a DP72 digital camera mounted on an Olympus BX53 light microscope. Chromosomal measurements were made using KaryoType software and karyotype formulae were determined. The karyotype formulae consisted of the following expressions: metacentric (m), submetacentric (sm), and acrocentric (a). The ideograms were drawn from the largest chromosome to the smallest chromosome based on the chromosomal lengths. The karyotype asymmetry was calculated according to the S/AI formula (symmetry/asymmetry index) (Eroğlu 2015).
COI sequence analysis
COI marker was used to generate sequences, as this marker has been widely used for species delimitation across various Parasitengona mites (Antonovskaia 2018; Young et al. 2012; 2019, 2021; Mąkol and Felska 2024). We generated COI sequences from four adults and a pool consisting of 20 larvae of T. latum.
DNA extraction was performed using the GeneJet Purification Kit (Thermo Fisher Scientific). A total PCR mixture of 50 μL was prepared with 17.5 μL of H₂O, 2.5 μL of each primer (10 pmol/μL), 25 μL of High Fidelity PCR Master Mix (Thermo Scientific), and 2.5 μL of DNA sample. The PCR conditions followed the protocol described by Hornok et al. (2015) using the LCO1490 and HCO2198 primers (Folmer et al. 1994). Samples were sent to Macrogen for sequencing. Sequences were aligned using the Clustal W program (Larkin et al. 2007) and compared using BLAST provided by the National Center for Biotechnology Information (NCBI) at http://blast.ncbi.nlm.nih.gov/Blast.cgi ![]() . The unique haplotype obtained in this study has been uploaded to GenBank (GenBank Accs. Number: PQ896991).
. The unique haplotype obtained in this study has been uploaded to GenBank (GenBank Accs. Number: PQ896991).
Results


Download as
Chromosome pair
Long arm length (μm)
Short arm length (μm)
Total length (μm)
Relative length (%)
Arm ratio
Centromeric index (%)
Chromosome type
1
1.17
0.75
1.92
23.10
1.56
39.06
metacentric
2
0.90
0.70
1.60
19.25
1.29
43.75
metacentric
3
0.83
0.52
1.35
16.25
1.60
38.52
metacentric
4
0.76
0.51
1.27
15.28
1.49
40.16
metacentric
5
0.91
0.21
1.12
13.48
4.33
18.75
acrocentric
6
0.82
0.23
1.05
12.64
3.56
21.90
acrocentric
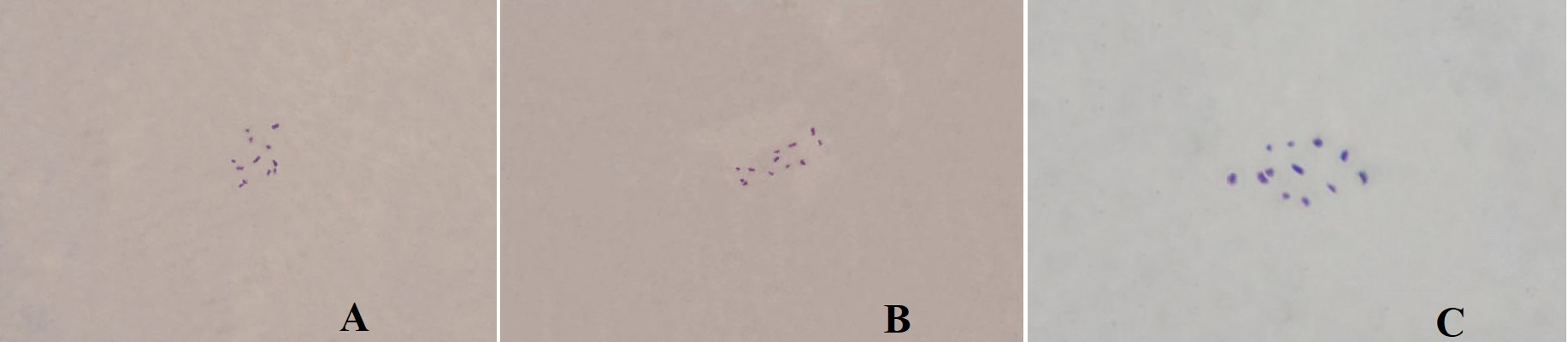
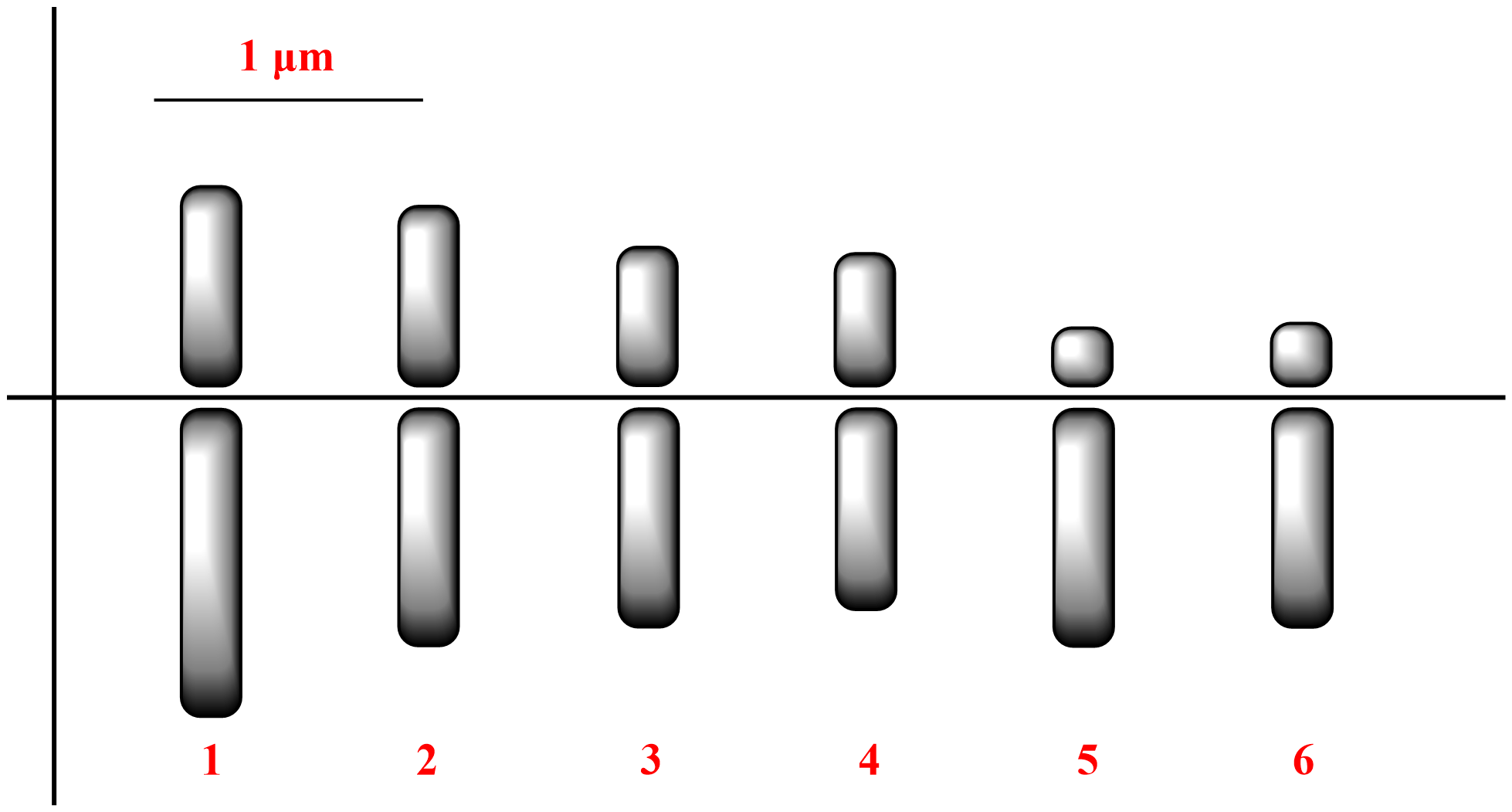
In T. latum, the diploid chromosome number was 2n=12 (Figure 1). The lengths of the small metacentric and acrocentric chromosomes ranged from 1.05 to 1.92 µm. The karyotype formula was 8m + 4a. The haploid chromosome length and mean chromosome length were 8.31 and 1.39 μm, respectively. The relative length and centromeric index ranged from 12.64 to 23.10 and from 18.75 to 43.75, respectively (Table 1). The S/AI value was 1.66. There was no satellite on the chromosomes (Figure 2).


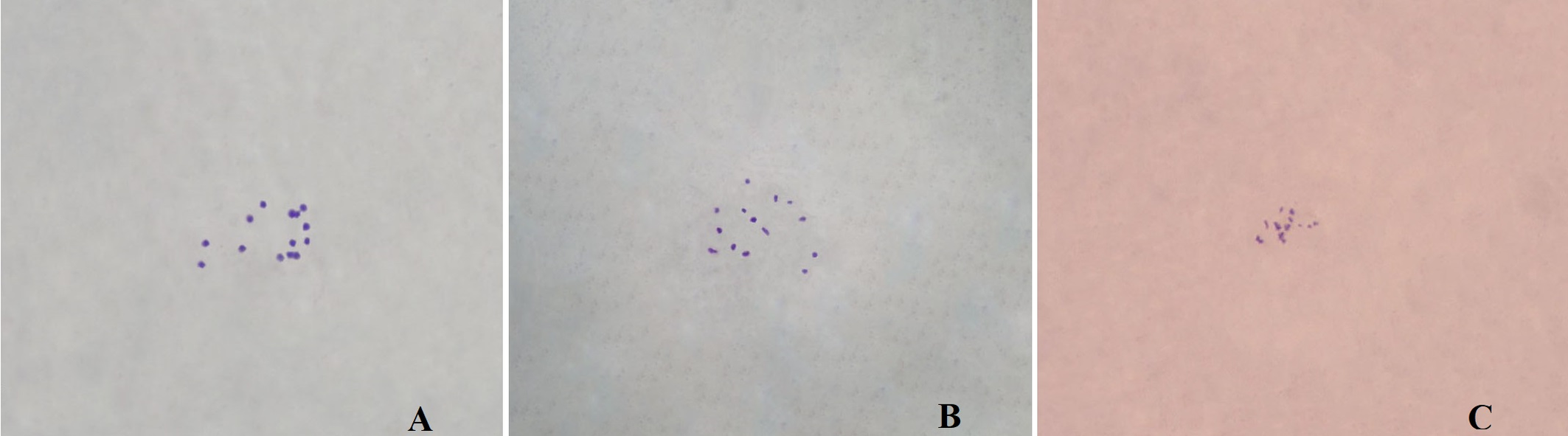
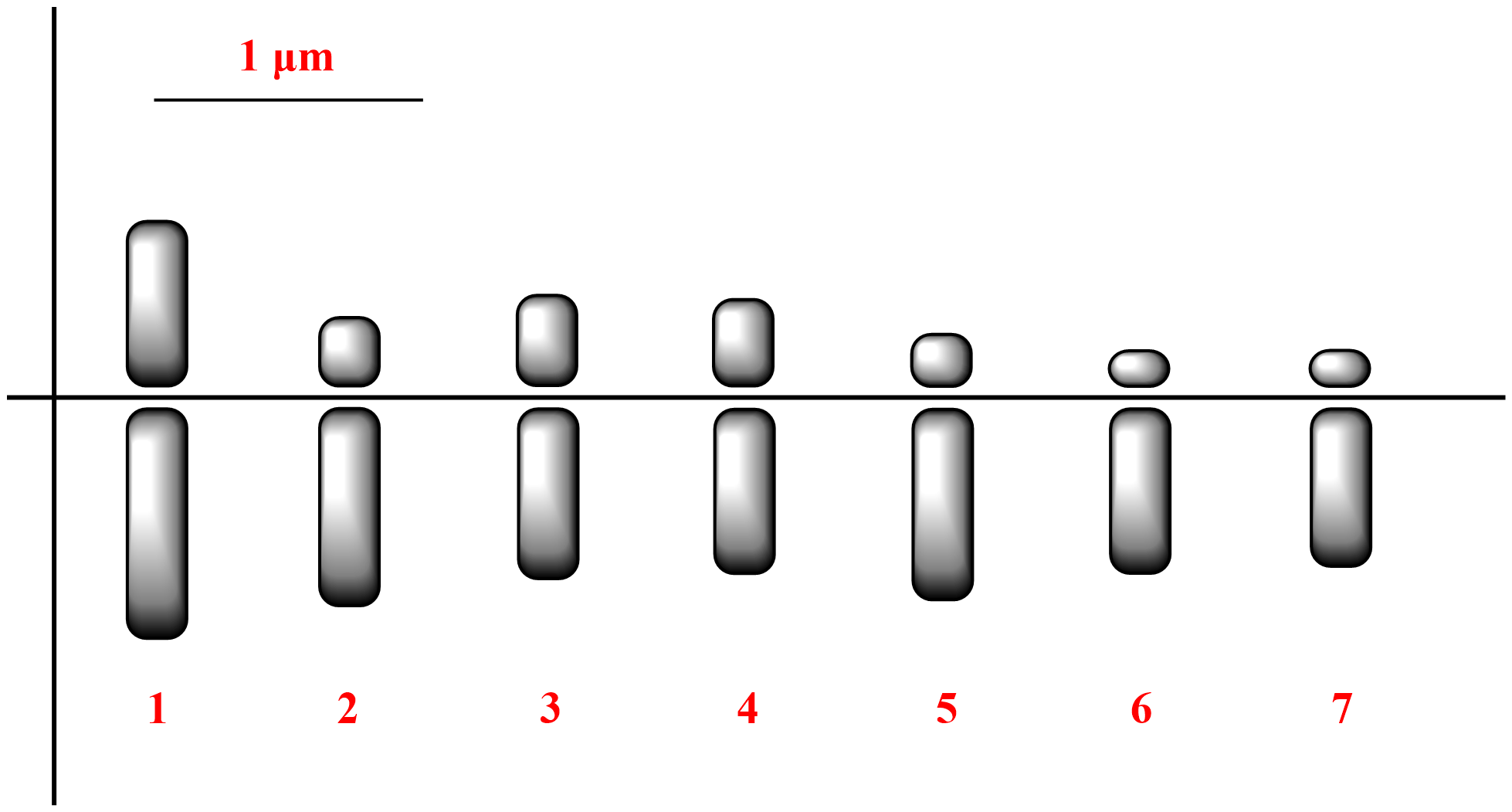
In E. trigonum, the diploid chromosome number was 2n=14 (Figure 3). The lengths of the small metacentric, submetacentric, and acrocentric chromosomes ranged from 0.68 to 1.40 µm. The karyotype formula was 2m + 6sm + 6a. The haploid chromosome length and mean chromosome length were 6.35 and 0.91 μm, respectively. The relative length and centromeric index ranged from 10.71 to 22.05 and from 18.57 to 42.14, respectively (Table 2). The S/AI value was 2.29. There was no satellite on the chromosomes (Figure 4).


Download as
Chromosome pair
Long arm length (μm)
Short arm length (μm)
Total length (μm)
Relative length (%)
Arm ratio
Centromeric index (%)
Chromosome type
1
0.81
0.59
1.40
22.05
1.37
42.14
Metacentric
2
0.68
0.25
0.93
14.65
2.72
26.88
Submetacentric
3
0.59
0.33
0.92
14.49
1.79
35.87
Submetacentric
4
0.56
0.31
0.87
13.70
1.80
35.63
Submetacentric
5
0.66
0.19
0.85
13.39
3.47
22.35
Acrocentric
6
0.57
0.13
0.70
11.02
4.38
18.57
Acrocentric
7
0.55
0.13
0.68
10.71
4.23
19.12
Acrocentric


COI sequence analysis and species identification
The comparison of COI sequences confirmed that the samples collected from Erzincan Province belong to T. latum. BLAST analysis showed 99.3% and 99.1% identity with the sequences PQ193923.1 and PQ193922.1, respectively, both derived from T. latum specimens collected in Poland (Mąkol and Felska 2024). The observed nucleotide differences (three and four base substitutions) therefore interpreted as geographic variation within the species rather than evidence of a distinct taxon.
Discussion
The selection of T. latum and E. trigonum in this study was initially based on their abundance at the sampling sites and their wide distribution in Europe (Mąkol and Wohltmann 2012), which suggests their ecological relevance. Notably, our study provides the first chromosomal data for both species, filling a significant gap in the cytogenetic knowledge of Trombidioidea. For T. latum, known issues of morphometric overlap with other congeners, especially in postlarval stages (Mąkol and Felska 2024), posed challenges for accurate identification. The integration of COI barcoding in our methodology proved essential for validating morphological diagnoses (Mąkol and Felska 2024), thereby strengthening the reliability of our karyological findings. In the case of E. trigonum, the species' distinct morphological characters facilitated identification in early developmental stages (Husband and Wohltmann 2011). By generating the first karyotypes for these taxa, our study offers a baseline for future comparative cytogenetic work and contributes to clarifying evolutionary and systematic relationships within Trombidiidae and Microtrombidiidae.
Download as * This species was reported by Onrat et al. (2006) as having 2n = 16 chromosomes. They referenced Sokolov (1954), but did not discuss the reason for the discrepancy in chromosome numbers.
Family
Genus
Species
Chromosome number
References
Erythraeidae
Erythraeus
Erythraus sp.
2n=16
Sokolov 1954
Trombiculidae
Leptotrombidium
L. akamushi
2n=12
Hoshiba et al. 2005
L. scutellare
2n=14
L. deliense
2n=14
Shirai et al. 1984
L. fletcheri
2n=14
L. arenicola
2n=28
Trombidiidae
Allothrombium
A. fuliginosum
2n=24
Sokolov 1954
Trombidium
T. latum
2n=12
NEW
Microtrombidiidae
Eutrombidium
E. trigonum
2n=14
Arrhenuridae
Arrhenurus
A. bicuspidator
2n=20
Sokolov 1954
A. papillator
2n=20
A. maculator
2n=10
A. pustulator
2n=26
A. (Megaluracarus) caudatus
2n=18
Eylaidae
Eylais
E. rimosa
2n=4
E. setosa
2n=4
E. mutila
2n=6
Hydrachnidae
Hydrachna
H. globosa
2n=12
H. leegei
2n=18
Hydrodromidae
Hydrodroma
H. despiciens*
2n=6
Sokolov 1954
Hydryphantidae
Hydryphantes
H. clypeatus
2n=6
H. bayeri
2n=10
Hydryhantes sp.
2n=10
H. ruber
2n=12
Thyas
T. dirempta
2n=18
Hygrobatidae
Hygrobates
H. calliger
2n=14
Lebertiidae
Lebertia
L. (Pilolebertia) porosa
2n=16
Frontipoda
F. musculus
2n=18
Limnesiidae
Limnesia
L. maculata
2n=18
L. undulata
2n=18
Limnocharidae
Limnochares
L. aquatica
2n=6
Pionidae
Piona
P. coccinea v. coccinea
2n=20
P. uncata
2n=20
P. carnea
2n=22
P. nodata
2n=8
Unionicolidae
Unionicola
U. crassipes
2n=18
Neumania
N. vernalis
2n=4
Eriophyidae
Aceria
A. sheldoni
2n=4
Helle and Wysoki 1983
Aculops
A. tetanothrix
2n=4
Aculus
A. persicae
2n=4
A. schlechtendali
2n=4
Artacris
A. macrorhynchus
2n=4
Phyllocoptruta
P. oleivora
2n=4
Phytoptus
P. tiliae
2n=4
Eupodidae
Linopodes
Linopodes sp.
2n=18
Sokolov 1954
Eupodes
Eupodes sp.
2n=18
Pyemotidae
Phytonemus
P. pallidus
2n=4
Bolland and Magowski 1995
P. fragariae
2n=4
Pyemotes
P. tritici
2n=6
Tarsonemus
T. confusus
2n=4
T. hermes
2n=4
T. nodosus
2n=4
Stigmaeidae
Agistemus
A. sanctiluciae
2n=4
Bolland and Ueckermann 1984
A. camerounensis
2n=4
Eupalopsellus
E. brevipalpus
2n=8
Eustigmaeus
E. bryonemus
2n=8
Flechtmann 1984
Tarsonemidae
Pediculopsis
P. graminum
2n=6
Zalewska 1979
P. ventricosus
2n=6
Polyphagotarsonemus
P. latus
2n=4
Flechtmann and Flechtmann 1984
Tenuipalpidae
Pentamerismus
P. taxi
2n=6
Helle and Wysoki 1983
Aegyptobia
Aegyptobia sp.
2n=4
Tetranychidae
Eotetranychus
E. uncatus
2n=6
Bolland et al. 1982
E. ranomofanae
2n=10
Zalewska 1979
E. carpini
2n=8
E. friedmanni
2n=6
E. sakalavensis
2n=4
Eutetranychus
E. africanus
2n=4
Bolland et al. 1982
Mononychellus
M. caribbeanae
2n=6
Gutierrez et al. 1991
Oligonychus
O. gramineus
2n=8
Bolland et al. 1982
O. leandrianae
2n=8
O. plegas
2n=8
O. thelytokus
2n=6
Panonychus
P. ulmi
2n=6
Bolland and Gotoh 1992
P. bambusicola
2n=6
P. thelytokus
2n=6
P. citri
2n=6
P. mori
2n=6
Petrobia
P. moutiai
2n=8
Bolland et al. 1982
Schizonobia
S. oudemansi
2n=8
Gutierrez and Bolland 1986
Schizotetranychus
S. reticulatus
2n=6
Bolland et al. 1982
Schizotetranychus sp.
2n=6
Helle and Wysoki 1983
Tetranychus
T. fijiensis
2n=8
Bolland et al. 1982
T. marianae
2n=8
T. lambi
2n=6
T. lombardinii
2n=6
T. macfarlanei
2n=6
T. yusti
2n=6
T. lintearius
2n=6
Tetranychus sp.
2n=10
Helle and Wysoki 1983
Tydeidae
Tydeus
T. caudatus
2n=4
Acaridida
Rhizoglyphus
R. echinopus
2n=10
Zalewska 1979
Tyrophagus
T. neiswanderi
2n=12
T. putrescentiae
2n=16
T. palmarum
2n=16
Acarus
A. siro
2n=18
Caloglyphus
C. berlesi
2n=18
C. michaeli
2n=18
Glycyphagus
G. domesticus
2n=18
Epidermoptidae
Myialges
M. pari
2n=16
Helle and Wysoki 1983
Gamasida
Amblyseius
A. arborens
2n=6
Wysoki and Bolland 1983
A. cucumeris
2n=8
A. potentillae
2n=8
A. isuki
2n=8
A. reductus
2n=8
A. salish
2n=8
Euseius
E. (A.) scutalis
2n=8
Iphiseius
I. degenerans
2n=8
Phytoseius
P. hawaiiensis
2n=8
Typhlodromus
T. (A.) transvaalensis
2n=8
T. annectens
2n=6
T. canadensis
2n=8
T. citri
2n=6
T. fallacis
2n=8
T. persianus
2n=8
T. porresi
2n=6
T. pyri
2n=8
Phytoseiidae
Typhlodromus
T. fallacis
2n=8
Zalewska 1979
T. occidentalis
2n=6
Argasidae
Argas
A. reflexus
2n=26
A. brumpti
2n=24
A. vespertilionis
2n=20
Ornithodorus
O. (Pavlovskyella) gurneyi
2n=12
Oliver and Bremner 1968
O. (Pavlovskyella) macmillani
2n=16
O. papillipes
2n=16
Sokolov 1954
O. qurneyi
2n=12
Zalewska 1979
O. tartakovskii
2n=26
Sokolov 1954
Ixodidae
Aponnoma
A. hydrosauri
2n=18
Zalewska 1979
Boophilus
B. calcaratus
2n=20
Sokolov 1954
B. microplus
2n=22
Garcia et al. 2002
Dermacentor
D. andersoni
2n=20
Oliver 1972
D. hunteri
2n=20
D. occidentalis
2n=20
D. parumapertus
2n=20
D. silvarum
2n=20
Sokolov 1954
D. variabilis
2n=20
Oliver 1972
Haemaphysalis
H. bispinosa
2n=20
Oliver et al. 1974
H. campanulata
2n=20
H. flava
2n=20
H. formosensis
2n=20
H. hystricis
2n=18
H. japonica
2n=20
H. kitaokai
2n=18
H. longicornis
2n=20
Oliver 1973
H. longicornis
2n=20
Oliver et al. 1974
H. megaspinosa
2n=20
H. pentalagi
2n=20
Ixodidae
Hyalomma
H. anatolicum
2n=20
Sokolov 1954
H. anatolicum excavatum
2n=20
H. asiaticum
2n=21
H. detritum
2n=21
H. dromedarii
2n=21
H. plumbeum (=marginatum)
2n=20
H. plumbeum plumbeum
2n=20
Ixodidae
Ixodes
I. ricinus
2n=28
Zalewska 1979
I. hexagonus
2n=26
I. holocyclus
2n=24
Ixodes spp.
2n=28
Kotsarenko et al. 2020
I. ricinus cell lines IRE/CTVM19
2n=23
I. ricinus cell lines IRE/CTVM20
2n=48
I. scapularis cell lines ISE18
2n=48
Rhipicaphalus
R. bursa
2n=20
Sokolov 1954
Opilioacaridae
Opilioacarus
O. thaleri
2n=16
Vazquez et al. 2021
Oribatida
Belba
B. verticillipes
2n=18
Sokolov 1954
Euzetes
E. semilunum
2n=18
Galumna
Galumna sp.
2n=18
Hypochthonius
H. rufulus
2n=18
Notaspis
N. punctatus
2n=18
Nothrus
N. palustris
2n=18
Zalewska 1979
N. silvestris
2n=18
Sokolov 1954
Brasilobates
B. spinosus
2n=16
Heethoff et al. 2006
B. spinosus
2n=16
Zhang et al. 2004
Galumna
G. longiporosa
2n=19
Heethoff et al. 2006
G. longiporosa
2n=19
Zhang et al. 2004
Hermannia
H. gibba
2n=16
Zalewska 1979
Hermanniella
H. gibber
2n=22
Gümüş et al. 2022
Hypochtonius
H. ryfulus
2n=18
Zalewska 1979
Notaspis
N. punctatus
2n=18
Oribotritia
O. hermanni
2n=14
Gümüş et al. 2022
Platynothorus
P. peltifer
2n=18
Zalewska 1979
Poroliodes
P. farinosus
2n=18
Trhypochthonius
T. tectorum
2n=18
Xenillus
X. tegeocranus
2n=18
Zygoribatula
Z. cognata
2n=30
Eroğlu and Per 2016
The karyotype data published to date are presented in Table 3. As illustrated in the table, there has been a gradual decline in interest in chromosomal studies over time. This decline may be attributed to the technical difficulties of cytogenetic analysis in mites and the greater focus on molecular methods in recent years. Diploid chromosome numbers in mites generally range from 4 to 30 (for detailed references, see Supplmeentary Table 1). The chromosome counts observed in T. latum and E. trigonum also fall within this range. Notably, as these represent the first chromosomal records for the families Trombidiidae and Microtrombidiidae, direct comparisons with closely related species are currently not possible. Nevertheless, the results presented here provide a valuable cytogenetic baseline and are expected to contribute significantly to the accumulation of karyological data within these understudied mite families. The S/AI parameter is widely used in detecting karyotype asymmetry (Eroğlu 2015). The karyotype of T. latum was ''symmetric'' type by S/AI value of 1.66 (1.0 < S/AI ≤ 2.0). The symmetrical karyotype indicates the early stages of karyotype evolution. The karyotype of E. trigonum was ''between symmetric and asymmetric'' type by S/AI value of 2.29 (2.0 < S/AI ≤ 3.0).
The phylogenetic utility of karyotype data in Trombidioidea remains difficult to assess due to the paucity of comparable chromosomal records within the group. While the diploid chromosome numbers and karyotype symmetry values presented here offer important baseline data for T. latum and E. trigonum, they cannot yet be reliably used for phylogenetic inference at higher taxonomic levels. Even in relatively better-studied mite groups, such as Ixodidae, significant intraspecific variation has been observed (e.g., Ixodes ricinus, Kotsarenko et al. 2020), suggesting that chromosomal characteristics may be subject to microevolutionary change. As such, the current variability within and across mite taxa cautions against overinterpretation of karyotype traits in the absence of broader comparative frameworks. Nonetheless, with future studies expanding cytogenetic datasets in mites, especially through standardized protocols, karyotype characters may eventually contribute to resolving phylogenetic relationships when combined with molecular and morphological data. The most important of these karyotype characters are basic and diploid chromosome numbers, chromosome type, and karyotype asymmetry. Especially chromosome type (monocentric or holocentric) constitutes an important differentiation model in many insect groups (Heethoff et al. 2006; Melters et al. 2012). While the chromosomes of T. latum and E. trigonum are monocentric, there are reports of holocentric chromosomes in mites (Eroğlu and Per 2016; Gümüş et al. 2018, 2022). However, monocentric chromosomes, which allow the determination of chromosome types, karyotype formula and karyotype asymmetry, are more prominent in terms of understanding karyotype evolution.
COI barcoding has emerged as a promising and increasingly valuable tool for accurately identifying mite species, particularly those within taxonomically challenging families like Trombidiidae. This molecular technique offers a standardized and complementary approach to traditional morphological identification, especially in morphologically cryptic groups. However, its effectiveness remains limited by the currently sparse sequence data available for many mite taxa, including Parasitengona. Only a limited number of species within the Trombidiidae family have reliable reference sequences available in public databases, which currently constrains the accuracy of molecular species delimitation (Young et al. 2019, 2021). In this study, COI barcoding successfully identified the specimens collected from Erzincan Province as T. latum, with 99.1–99.3% sequence identity to reference sequences from Poland. These values fall within the widely accepted threshold for intraspecific variation (≤2%) for species-level identification (Hebert et al. 2003; Young et al. 2019, 2021), supporting conspecificity despite three to four nucleotide differences. These minor variations likely reflect geographic differentiation within the species. Our results not only confirm the identity of T. latum through molecular means but also contribute to closing the molecular data gap in Parasitengona, underscoring the need for further studies with broader geographic sampling and additional genetic markers to explore population structure and evolutionary relationships in this understudied group.
While our results provide preliminary cytogenetic and molecular data for two trombidioid mite species, further integrative studies will help to more fully evaluate the potential of combining these approaches for species delimitation. The current findings offer a valuable reference point; however, certain limitations should be acknowledged. Chromosomal analyses were based on a limited number of individuals, and further studies are needed to assess intraspecific karyotypic variability. Likewise, although COI barcoding was informative for species identification, reliance on a single gene and a limited reference dataset restricts broader phylogenetic inference. In addition, we acknowledge that many of the available COI reference sequences are tied to species hypotheses that were originally based on limited morphological datasets, often involving a small number of specimens from single localities. In Parasitengona, intraspecific variation in morphological traits is rarely assessed, and other informative criteria such as reproductive isolation or life cycle data are largely absent. Consequently, COI comparisons may reflect similarity to potentially unstable or insufficiently defined species concepts. While we interpret our results as strong support for the identification of T. latum, we also recognize that molecular species delimitation in mites—especially in understudied groups—remains tentative unless supported by integrative evidence. This reinforces the importance of developing broader reference frameworks and expanding comparative analyses that combine molecular, morphological, and ecological data (Blaxter 2004; Dayrat 2005; Knowlton 2000). Future research incorporating multi-locus approaches and comparative karyotype analyses across related taxa will be critical for advancing our understanding of mite evolution and taxonomy.
Acknowledgments
This study is part of the master's thesis of the first author and was presented as a short summary (only T. latum) at 3rd International Symposium on Biodiversity Research 20-22 October 2021, Erzurum, Türkiye. This study was financially supported by the Scientific Research Fund of Erzincan Binali Yıldırım University (EBYU), research project number FYL-2020-748. We are very grateful to EBYU for its financial support. We would like to thank Alper Torunlar for his assistance in collecting T. latum specimens during fieldwork, and Nisa Gümüş for her help with the laboratory work. Also, we are very grateful to the two anonymous referees for their valuable comments, which also contributed significantly to the improvement of the manuscript.
References
- Antonovskaia A.A. 2018. Using DNA markers in studies of chigger mites (Acariformes, Trombiculidae). Entomol. Rev., 98: 1351-1368. https://doi.org/10.1134/S0013873818090130
- Blaxter M. 2004. The promise of a DNA taxonomy. Philos. Trans. R. Soc. B: Biol. Sci., 359(1444): 669-679. https://doi.org/10.1098/rstb.2003.1447
- Bolland H., Gutierrez J., Helle W. 1982. Chromosomes in spider mites (Tetranychidae- Acari). Acarologia, 23(3): 271-275.
- Bolland H.R., Gotoh T. 1992. Chromosome numbers in the genus Panonychus in Japan (Acari: Tetranychidae). Appl. Entomol. Zool., 27(4): 602-604. https://doi.org/10.1303/aez.27.602
- Bolland H.R., Magowski W.L. 1995. The chromosome number in tarsonemid and pyemotid mites (Acari: Heterostigmata: Tarsonemidae, Pyemotidae). Entomol. Ber., 55(10): 166-167.
- Bolland H.R., Ueckermann E.A. 1984. Raphignathoid mites (Acari: Prostigmata) from Cameroun with reference to their chromosome numbers. Phytophylactica, 16(3): 201-108. https://doi.org/10.10520/AJA03701263_871
- Cooper K.W. 1937. Reproductive behavior and haploid parthenogenesis in the grass mite, Pediculopsis graminum. Proc. Natl. Acad. Sci. U.S.A., 23: 41-44. https://doi.org/10.1073/pnas.23.2.41
- Cooper K.W. 1939. The nuclear cytology of the grass mite, Pediculopsis graminum, with special reference to karyomerokinesis. Chromosoma, 1: 51- 103. https://doi.org/10.1007/BF01271623
- Dayrat B. 2005. Toward integrative taxonomy. Biol. J. Linn. Soc., 85(3): 407-415. https://doi.org/10.1111/j.1095-8312.2005.00503.x
- Eroğlu H.E. 2015. Which chromosomes are subtelocentric or acrocentric? A new karyotype symmetry/ asymmetry index. Caryologia, 68: 239-245. https://doi.org/10.1080/00087114.2015.1032614
- Eroğlu H.E., Per S. 2016. Karyotype analysis of Zygoribatula cognata (Oudemans) (Acari: Oribatida: Oribatulidae). Turk. J. Entomol., 40(1): 33-38. https://doi.org/10.16970/ted.50556
- Flechtmann C.H.W. 1984. Eustigmaeus bryonemus, sp. n., a moss-feeding mite from Brasil (Acari, Prostigmata: Stigmaeidae). Rev Brasil. Zool., 2: 387-391. https://doi.org/10.1590/S0101-81751984000200010
- Flechtmann C.H.W., Flechtmann C.A.H. 1984. Reproduction and chromosomes in the broad mite, Polyphagotarsonemus latus (Bank,1904) (Acar, Prostigmata, Tarsonemidae), pp. 455-456. In: Griffith D.A., Bowman C., Acarology, VI, vol. 1(1), Ellis Horwood Ltd., Chichester.
- Folmer O., Black M., Hoeh W., Lutz R., Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3: 294-299.
- Gabryś G. 1999. The world genera of Microtrombidiidae (Acari, Actinedida, Trombidioidea).
- Monogr. Uppe. Siles. Mus. 2: 1-361.
- Garcia R.N., Garcia-Fernandez C., Garcia S.M.L., Valente V.L.S. 2002. Mitotic and meiotic chromosomes of a southern Brazilian population of Boophilus microplus (Acari, Ixodidae). Iheringia Ser. Zool., 92: 51-57. https://doi.org/10.1590/S0073-47212002000100004
- Gokhman V.E., Quicke D.L.J. 1995. The last twenty years of parasitic Hymenoptera karyology: An update and phylogenetic implications. J. Hymenopt. Res., 4: 41-63.
- Gutierrez J., Bolland H.R. 1986. Description and karyotype of Schizonobia oudemansi n. sp. from The Netherlands (Acari: Tetranychidae). Entomol. Ber., 46(1): 39-43.
- Gutierrez J., Bolland H.R., Etienne J., Cotton D. 1991. Deuterotoky in Mononychellus caribbeanae (Acari: Tetranychidae) on Cassava in Guadeloupe. Karyotype and preliminary biological data. Modern Acarology, vol. 2: 443-447. https://hal.inrae.fr/hal-02851556
- Gümüş N., Per S. Eroğlu H.E. 2018. Karyotype analysis of Phauloppia lucorum (Koch, 1841) (Oribatida: Oribatulidae). Turk. J. Entemol., 42, 77-83. https://doi.org/10.16970/entoted.382980
- Gümüş N., Eroğlu H.E., Per S. 2022. Karyotype analysis of two oribatid mite species (Acari: Oribatida). Acarol. Stud., 4(1): 41-45. https://doi.org/10.47121/acarolstud.1059305
- Hansell R.I.C., Mollison M.M., Putman W.L. 1964. A cytological demonstration of arrhenotoky in three mites of the family Phytoseiidae. Chromosoma, 15: 562-67. https://doi.org/10.1007/BF00319990
- Hebert P.D.N., Ratnasingham S., DeWaard J.R. 2003. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. Lond. Series B: Biol. Sci., 270 (Suppl.1): 96-99. https://doi.org/10.1098/rsbl.2003.0025
- Heethoff M., Bergmann P., Norton R.A. 2006. Karyology and sex determination of oribatid mites. Acarologia, 46(1-2): 127-131.
- Helle W., Bolland H.R. 1967. Karyotypes and sex-determination in spider mites (Tetranychidae). Genetica, 38: 43-53. https://doi.org/10.1007/BF01507446
- Helle W., Wysoki M. 1983. The chromosomes and sex-determination of some actinotrichid taxa (Acari), with special reference to Eriophyidae. Int. J. Acarol., 9(2): 67-71. https://doi.org/10.1080/01647958308683315
- Hornok S., Estrada-Pe˜na A., Kontsch'an J., Plantard O., Kunz B., Mihalca A.-D., Thabah A., Tomanovi'c S., Burazerovi'c J., Tak'acs N. 2015. High degree of mitochondrial gene heterogeneity in the bat tick species Ixodes vespertilionis, I. ariadnae and I. simplex from Eurasia. Parasit. Vectors, 8: 1-8. https://doi.org/10.1186/s13071-015-1056-2
- Hoshiba H., Takahashi M., Misumi H., Urakami H. 2005. Chromosome studies of Leptotrombidium akamushi and L. scutellare (Acari: Trombiculidae) in Japan. Int. J. Acarol., 31(2): 171-174. https://doi.org/10.1080/01647950508683669
- Husband R.W., Wohltmann A. 2011. A redescription of Eutrombidium locustarum (Walsh) (Acari: Microtrombidiidae) and a new North American Podapolipoides (Acari: Podapolipidae), parasites of Schistocerca piceifrons (Walker) (Orthoptera: Acrididae) from Yucatan, Mexico. Int. J. Acarol., 37: 260-292. https://doi.org/10.1080/01647954.2011.564592
- Imai H., Taylor R.W. Crosland M.V., Crozier R. 1988. Modes of spontaneous chromosomal mutation and karyotype evolution in ants with reference to the minimum interaction hypothesis. Jpn. J. Genet., 63(2): 159-185. https://doi.org/10.1266/jjg.63.159
- Knowlton N. 2000. Molecular genetic analyses of species boundaries in the sea. Hydrobiologia, 420(1): 73-90. https://doi.org/10.1023/A:1003933603879
- Kotsarenko K., Vechtova P., Lieskovska J., Füssy Z., Cabral-de-Mello D.C., Rego R.O., Alberdi P., Collins M., Bell-Sakyi L., Sterba J., Grubhoffer L. 2020. Karyotype changes in long-term cultured tick cell lines. Sci. Rep., 10(1): 13443. https://doi.org/10.1038/s41598-020-70330-5
- Larkin M.-A., Blackshields G., Brown N.-P., Chenna R., McGettigan P.-A., McWilliam H., Valentin F., Wallace I.-M., Wilm A., Lopez R., Thompson J.-D. 2007. Clustal W and Clustal X version 2.0. Bioinformatics, 23(21): 2947-2948. https://doi.org/10.1093/bioinformatics/btm404
- Mąkol J. 2002. A redescription of Trombidium latum (C. L. Koch, 1837) (Acari: Actinotrichida, Trombidioidea) with characteristics of all active instars. Ann. Zool., 52: 433-442.
- Mąkol J. 2005. Trombidiidae (Acari: Actinotrichida: Trombidioidea) of Poland. Fauna Poloniae, Museum and Institute of Zoology, Polish Academy of Sciences & Natura Optima Dux Foundation, Warsaw, Vol. 1 [NS]: 1-259.
- Mąkol J., Sevsay S. 2011. Notes on the genus Dolichothrombium (Acari: Prostigmata: Trombidiidae) with description of a new species. Zootaxa, 2971: 1-16. https://doi.org/10.11646/zootaxa.2971.1.1
- Mąkol J., Felska M. 2024. The syntopic occurrence of velvet mites Trombidium spp. (Acariformes: Trombidiidae) with the first characteristics of the larva of Trombidium heterotrichum. Sci. Rep., 14: 32155. https://doi.org/10.1038/s41598-024-81750-y
- Mąkol J., Wohltmann A. 2012. An annotated checklist of terrestrial Parasitengona (Actinotrichida: Prostigmata) of the World, excluding Trombiculidae and Walchiidae. Ann. Zool., 62(3): 359-562. https://doi.org/10.3161/000345412X656671
- Mąkol J., Wohltmann A. 2000. A redescription of Trombidium holosericeum (Linnaeus, 1758) (Acari: Actinotrichida, Trombidioidea) with characteristics of all active instars and notes on taxonomy and biology. Ann. Zool., 50: 67-91.
- Melters D.P., Paliulis L.V., Korf I.F., Chan S.W. 2012. Holocentric chromosomes: convergent evolution, mei-otic adaptations, and genomic analysis. Chromosome Res., 20 (5): 579-593. https://doi.org/10.1007/s10577-012-9292-1
- Norton R.A., Kethley J.B., Johnston D.E., OConnor B.M. 1993. Phylogenetic perspectives on genetic systems and reproductive modes of mites, pp. 8-99. In: Evolution and diversity of sex ratio in insects and mites. In: Wrensch D. and Ebbert, M.A. (Eds). Chapman and Hall, New York, USA, 1993. https://doi.org/10.1007/978-1-4684-1402-8_2
- Oliver J.H. 1964. Comments on karyotypes and sex determination in the Acari. Acarologia, 6: 288-93.
- Oliver J.H. 1965. Cytogenetics of ticks. 2. Multiple sex chromosomes. Chromosoma, 17: 323-27. https://doi.org/10.1007/BF00283388
- Oliver J.H. 1972. Cytogenetics of ticks (Acari: Ixodoidea) 6. Chromosomes of Dermacentor species in the United States. J. Med. Entomol., 9(2): 177-182. https://doi.org/10.1093/jmedent/9.2.177
- Oliver J.H. 1973. Cytogenetics of Ticks (Acari: Ixodoidea) 12. Chromosome and hybridization studies of bisexual and parthenogenetic Haemaphysalis longicornis races from Japan and Korea. Chromosoma, 42: 269-288. https://doi.org/10.1007/BF00284775
- Oliver J.H. 1977. Cytogenetics of mites and ticks. Annu. Rev. Entomol., 22: 407-429. https://doi.org/10.1146/annurev.en.22.010177.002203
- Oliver J.R J.H., Bremner K.C. 1968. Cytogenetics of ticks. III. Chromosomes and sex determination in some Australian hard ticks (Ixodidae). Annals of the Entomological Society of America, 61(4): 837-844. https://doi.org/10.1093/aesa/61.4.837
- Oliver J.H., Tanaka K., Sawada M. 1973. Cytogenetics of ticks. 12. Chromosomes and hybridization studies of bisexual and parthenogenetic Haemaphysalis longicornis races from Japan and Korea. Chromosoma, 42: 269-88. https://doi.org/10.1007/BF00284775
- Oliver J.H., Tanaka K., Sawada M. 1974. Cytogenetics of Ticks (Acari: Ixodoidea) 14. Chromosomes of nine species of Asian Haemaphysalines. Chromosoma, 45: 445-456. https://doi.org/10.1007/BF00283388
- Onrat S.T., Aşçı F., Özkan M. 2006. A cytogenetics study of Hydrodroma despiciens (Müller, 1776) (Acari: Hydrachnellae: Hydrodromidae). Genet. Mol. Res., 5(2): 342-349.
- Patau K. 1934. Cytologische Untersuchungen an der haploparthenogenetischen. Milbe Pediculoides ventricosus. Z. Jahrb., Allg. Zool., Bd. 56.
- Reuter E. 1909. Ferokinesis, ein neur Kernteilungsmodus. Acta Soc. Sci. Fenn, 36: 4.
- Robaux P. 1967. Contribution à l'étude des Acariens Thrombidiidae d'Europe: 1 Étude des Thrombidions adultes de la Péninsule Iberique 2 Liste critique des Thrombidions d'Europe. Mémoires du Muséum National d'Histoire Naturelle, 46: 1-124.
- Schrader F. 1923. Haploidy bei einer Spinnenmilbe. Arch. Midrosk. Anat, 97: 610-22. https://doi.org/10.1007/BF02977822
- Shirai A., Ram S., Gan E., Lewis J.-R., Kanda T., Chiang G.L., Groves M.G. 1984. Comparative studies on the karyotypes of Leptotrombidium deliense, L. fletcheri, and L. arenicola (Acari: Trombiculidae). J. Med. Entomol., 21(5): 616-617. https://doi.org/10.1093/jmedent/21.5.616
- Sokolov I.I. 1934. Untersuchungen uber die Spermatogenese bei den Arachniden. V. Uber die Spermatogenese der Parasitidae. Z. Zellforsch, 21: 42-109. https://doi.org/10.1007/BF00668270
- Sokolov I.I. 1945. Karyological study of some Acari and the problem of sex determination in the group. Izv. Akad. Nauk, 6: 654-63.
- Sokolov I.I. 1954. The chromosome complex of mites and its importance for systematics and phylogeny. Trud. Leningr. Obshch. Estestvois., 72: 124-59.
- Vázquez M. M., Herrera I.M.Á., Just P., Lerma A.C.R., Chatzaki M., Heller T.L., Král J. 2021. A new opilioacarid species (Parasitiformes: Opilioacarida) from Crete (Greece) with notes on its karyotype. Acarologia, 61(3): 548-563. https://doi.org/10.24349/acarologia/20214449
- Walsh B.D. 1866. Answers to corresponds: Walter Riddel, Canada West. The Practical Entomologist, 1(12): 126.
- Warren E. 1940. On the genital system of Dermanyssus gallinae and several other Gamasidae. Ann. Natal. Mus., 9: 409-59.
- Wohltmann A., Wendt F.E. 1996. Observations on the biology of two hygrobiotic trombidioid mites (Acari: Prostigmata: Parasitengonae), with special regard to host recognition and parasitism tactics. Acarologia, 37(1): 31-44.
- Wysoki M. 1973. Further studies on karyotypes and sex determination of phytoseiid mites. Genetica, 44: 139-45. https://doi.org/10.1007/BF00155967
- Wysoki M., Bolland H.R. 1983. Chromosome studies of phytoseiid mites (Acari: Gamasida). Int. J. Acarol., 9(2): 91-94. https://doi.org/10.1080/01647958308683319
- Wysoki M., Swirski E. 1968. Karyotypes and sex determination of ten species of phytoseiid mites. Genetica, 39: 220-28. https://doi.org/10.1007/BF02324464
- Young M.R., Behan-Pelletier V.M., Hebert P.D. 2012. Revealing the hyperdiverse mite fauna of subarctic Canada through DNA barcoding. PLoS One, 7(11): e48755. https://doi.org/10.1371/journal.pone.0048755
- Young M.R., DeWaard J.R., Hebert P.D. 2021. DNA barcodes enable higher taxonomic assignments in the Acari. Sci. Rep., 11(1): 15922. https://doi.org/10.1038/s41598-021-95147-8
- Young M.R., Proctor H.C., Dewaard J.R., Hebert P.D. 2019. DNA barcodes expose unexpected diversity in Canadian mites. Mol. Ecol., 28(24): 5347-5359. https://doi.org/10.1111/mec.15292
- Zalewska M. 1979. Kariosystematyka roztoczy. Przegl. Zool., 23: 140-148.
- Zhang P.J., Fu R.S., Wang Y.H. 2004. Karyotype analysis of two species of Oribatida (Acari). Acarologia, 44(3-4): 231-234.



2025-04-11
Date accepted:
2025-06-17
Date published:
2025-06-25
Edited by:
Mąkol, Joanna

This work is licensed under a Creative Commons Attribution 4.0 International License
2025 Karağaç, Rümeysa; Eroğlu, Halil Erhan; Hekimoğlu, Olcay and Sevsay, Sevgi
Download the citation
RIS with abstract
(Zotero, Endnote, Reference Manager, ProCite, RefWorks, Mendeley)
RIS without abstract
BIB
(Zotero, BibTeX)
TXT
(PubMed, Txt)



