Genetic and morphological features of some Turkish populations of Amblyseius swirskii (Athias-Henriot) (Acari: Phytoseiidae)
Mertoglu Boz, Gamze  1
; Tixier, Marie-Stéphane
1
; Tixier, Marie-Stéphane  2
; Douin, Martial
2
; Douin, Martial  3
and Kumral, Nabi Alper
3
and Kumral, Nabi Alper  4
4
1✉ Department of Plant Protection, Faculty of Agriculture, Uludag University, Gorukle Campus, Bursa, 16059, Türkiye.
2CBGP, Institut Agro Montpellier, INRAE, CIRAD, IRD, Univ. Montpellier, Montpellier, France.
3CBGP, Institut Agro Montpellier, INRAE, CIRAD, IRD, Univ. Montpellier, Montpellier, France.
4Department of Plant Protection, Faculty of Agriculture, Uludag University, Gorukle Campus, Bursa, 16059, Türkiye.
2024 - Volume: 64 Issue: 4 pages: 1163-1174
https://doi.org/10.24349/la7w-80psOriginal research
Keywords
Abstract
Introduction
Predatory mites from the family Phytoseiidae (Acari: Mesostigmata) play a crucial role in biological control (McMurtry et al. 2013; Tixier 2018). Various species are used to control pest mites and small insect in many crops (Kostiainen and Hoy 1996; McMurtry and Croft 1997). Phytoseiid mites are reported on every continent except Antarctica (Tixier et al. 2008a; McMurtry et al. 2013), with more than 2,500 valid species described to date (Chant and McMurtry 2007; Demite et al. 2024). Several studies have investigated intraspecific variations in some phytoseiid species for taxonomy and biological adaptation. Morphological and molecular analyses have revealed significant intraspecific variations, which are critical for accurate species identification (Okassa et al. 2011; Tixier et al. 2017; Lima et al. 2018; Tixier et al. 2021). These variations are also associated to ecological factors such as host plants, climatic conditions, seasonal changes (i.e., overwintering females), and prey species. For instance, Tixier et al. (2010a), demonstrated genetic differentiation between populations of Phytoseiulus longipes Evans, based on their ability to feed or not on Tetranychus evansi Baker & Pritchard. Similarly, Queiroz et al. (2021) and Tixier et al. (2022a) suggested that genetic differentiation in populations of Phytoseiulus macropilis Banks, and several other phytoseiid species - such as Amblyseius swirskii Athias‐Henriot, Kampimodromus aberrans (Oudemans), Neoseiulus californicus (McGregor), Phytoseius finitimus Ribaga, Phytoseiulus persimilis Athias‐Henriot, Typhlodromus (Typhlodromus) pyri Scheuten, and Typhlodromus (Anthoseius) recki Wainstein - might be associated to climatic conditions. Tixier et al. (2003, 2020) also showed morphological differentiation in T. (A.) recki and K. aberrans based on their host plants. This study focuses on A. swirskii, one of the most widely used phytoseiid species alongside with N. californicus and P. persimilis (Knapp et al. 2018). This species preys on tetranychid and eriophyid mites, thrips, whiteflies, and pollen (Lee and Gillespie 2011; McMurtry et al. 2013; Buitenhuis et al. 2015). This predatory mite is used in various crops, including greenhouses, vineyards, fruit orchards, vegetable fields, and citrus groves (Castagnoli and Falchini 1993). It is commercially available in Türkiye since 2009, and is also known to occur naturally in this country (Döker and Kazak 2019; Demite et al. 2024). Notably, it was the the predominant phytoseiid species found in commercial citrus orchards in Adana province (Southern Türkiye) between 2009 and 2015 (Kazak et al. 2016). Tixier et al. (2022b) recently studied molecular variations in this species across Europe and Africa, including populations commercialized by several companies. They showed very low genetic variation among these populations, with the exception of the population from Cape Verde. They proposed two hypotheses: (i) low intrinsic genetic variation within the species, or (ii) an overdispersion of commercial populations replacing the natural ones. Turkish populations of A. swirskiii were not included in their study, despite the species' naturally occurrence in this country on various crops. The present study aims to investigate the genetic variations in Turkish populations of A. swirskii and compare their DNA sequences with those of commercial populations to test these latter hypotheses. Furthermore, because morphological traits can also reflect ecological adaptations, this study includes morphological analyses to provide additional elements for taxonomical purposes and species identification.
Material and methods
Populations considered
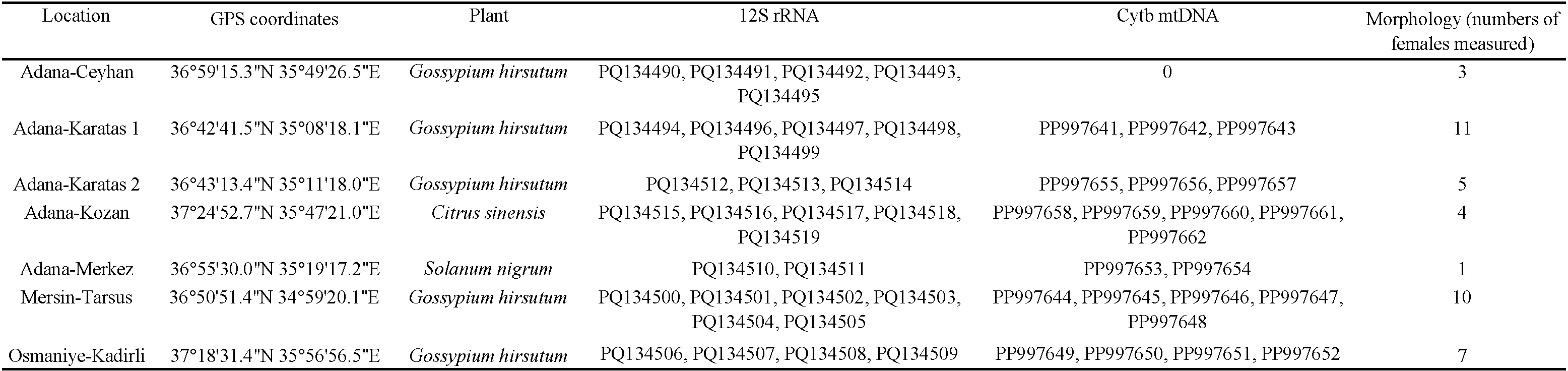


Specimens of A. swirskii were collected from crops (cotton: Gossypium hirsutum L., Malvaceae and orange: Citrus sinensis (L.) Osbeck, Rutaceae) and a weed species (Solanum nigrum L., Solanaceae) in different locations in 2022 and 2023. Collection sites include various locations in Adana (Ceyhan, Karataş 1, Karataş 2, Kozan, Merkez), as well as in Mersin (Tarsus) and Osmaniye (Kadirli) provinces (Supplementary file 1). Samples were obtained from commercial cotton and orange orchards, where A. swirskii had not been released. Additionally, specimens were collected on S. nigrum in one orchard. Table (1) shows details on the localities, the plants sampled, and the numbers of specimens considered per population for both molecular and morphological analyses. Although the specimens for morphological and molecular analyses were sourced from the same populations, they were not exactly the same specimens due to the measurement distortions during DNA extraction.
Morphological analyses
Forty-one A. swirskii females from seven populations were mounted on slides in Hoyer's medium and observed using a phase and differential interference contrast microscope (Leica DMLB, Leica Microsystèmes, Le Pecq, France) (40× magnification). In phytoseiid mite taxonomy, the length of dorsal setae is commonly considered important. Therefore, measurements of 25 dorsal idiosomal setae of females were obtained: j1, j3, j4, j5, j6, J2, J5, z2, z4, z5, Z1, Z4, Z5, s4, S2, S4, and S5. Additionally, macrosetae on the leg IV (SgeIV, StiIV, STIV) and the dimensions (length and width at level of z4) of the dorsal shield (DSL and DSW, respectively), as well as length and the width of the ventrianal shield (at level of ZV2 and at level of anus), were measured. The idiosomal setal terminology used in this study follows that of Lindquist and Evans (1965), as adapted by Rowell et al. (1978) for the family Phytoseiidae. The terminology of macrosetae on the leg IV follows the terminology of Athias-Henriot (1957). All measurements are provided in micrometers. As for one population only one specimen was available, ANOVA analyses were not performed. Instead, a Principal Component Analysis (PCA) was conducted to determine if the combination of morphological characters could enable differentiate among the specimens considered. The statistical analyses were conducted using R Core Team (2021) software, with the ''FactoMineR'' (Lê et al. 2008) and ''factoextra'' packages (Kassambara and Mundt 2020).
Molecular analyses
Total DNA was extracted from each specimen using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany), following the ′Purification of Total DNA from Animal Blood or Cells' protocol (Spin-Column Protocol), modified for extracting total DNA from mites (Kanouh et al. 2010a, b; Tixier et al. 2010a). Following DNA extraction, the mite carcasses were collected and mounted on slides using the procedure described by Tixier et al. (2010b) to confirm species identity.
The primers used to amplify the DNA fragments were: 12S rRNA, 5′-3′ TACTATGTTACGAC TTAT ve 3′-5′ AAACTAGGATTAGATACCC (Jeyaprakash and Hoy 2002), Cytb mtDNA, 5′–3′ TAWRAARTATCAYTCDGGTTKRATATG, and 3′–5′ CCWTGAGGACAAATAWSWTTYTGAGG (Tixier et al. 2012). Polymerase chain reactions (PCR) were performed in a 25 μL volume, containing 4 μL of mite DNA, 2.5 μL (1 mM) of buffer X, 1 μL (1.5 mM) of MgCl2, 0.5 μL (0.05 mM for each) DNTPs, 0.175 mL (0.7 mM) for each primer, 0.125 mL (0.625 U) of Taq Polymerase (Qiagen) and 16.525 μL of water (Tixier et al. 2012). Thermal cycling conditions were as follows, for the 12S rRNA marker: 95 °C for 1 min, followed by 35 cycles of 94 °C for 30 sec, 40 °C for 30 sec and 72 °C for 1 min, and 72 °C for 5 min. For the Cytb mtDNA marker: 94 °C for 3 min, followed by 35 cycles of 92 °C for 20 sec, 53 °C for 1 min, and 72 °C for 1 min and 72 °C for 5 min (Tixier et al. 2012). PCR products were sequenced along both strands using Dynamic ET Terminator Cycle Sequencing kit, and purified using ExoSAP-IT (Amersham). Sequencing was performed using the Megabase 1,000 apparatus. The sequences were analysed on both strands (forward and reverse), and the resulting consensus sequences were compared to those in the NCBI GenBank database to identify potential contaminations. Sequences alignment and analyses were performed using the MEGA X software (Kumar et al. 2018). Genetic distances were calculated using the Kimura-2-parameter model, and maximum likelihood trees were constructed using the substitution of evolution models TrN+I for the 12S rRNA sequences and GTR+I for the Cytb mtDNA sequences. Node support was determined using 1000 bootstrap replicates. Amblyseius andersoni (Chant) was selected as the outgroup species, as it belongs to the same sub-family (Amblyseiinae) and is morphologically similar to A. swirskii (12S rRNA and CytB mtDNA GenBank assession numbers: HQ404858, KU318207). The GenBank accessions numbers for the 12S rRNA sequences of specimens commercialized by Agrobio, Koppert, and Bioplanet are MW404026, MW404023 and MW404028, respectively. The GenBank accessions numbers for the Cytb mtDNA sequences of specimens commercialized by Koppert are MT828779, MT828780, and MT828781.
Results
Morphological analyses
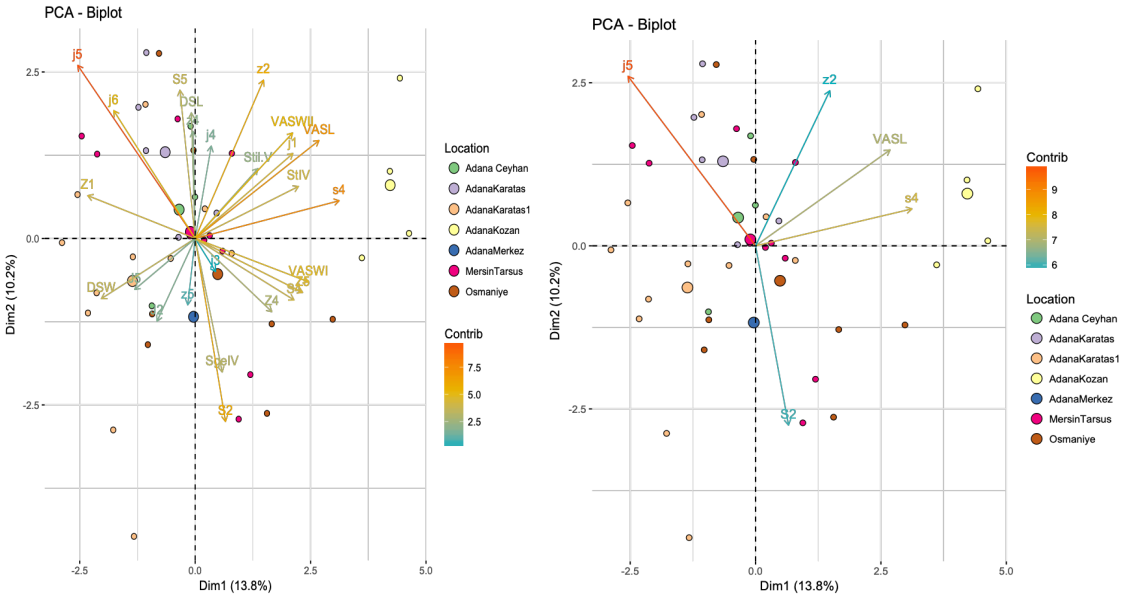


The supplementary file (2) shows the means and min-max values of the 25 morphological traits of the seven populations studied. The PCA (Principal Component Analysis) explains 24% of the total variation of the system, with axis 1 explaining for 13.8% and axis 2 for 10.2% (Figure 1). The five most well represented morphological traits were the length of the setae j5, z2, s4, S2, and the ventrianal shield (VAS). Generally, specimens from the same population were not grouped together, except for those collected from orange trees in Adana Kozan and, to a lesser extent, the populations from Adana Karatas and Adana Karatas 2. Specifically: (i) the population collected on orange in Adana Kozan was characterized by shorter seta j5, and longer seta s4 and of VAS, (ii) the Adana Karatas population on cotton was characterized by longer setae j5 and z2, with shorter seta S2, and (iii) the Adana Karatas 2 population differs from Adana Karatas 1, especially along axis 2, which is mainly explained by lengths of setae j5 and S2. No clear morphological differentiation among populations based on geographic location was observed. When PCA was performed based on the plant species from which specimens were collected (Figure 2), specimens on orange trees were distinctly separated from those collected on cotton. A great variation was observed among specimens collected from cotton across various locations. The specimens collected on orange trees came from a single population, which may explain their homogeneity and distinct separation from the cotton specimens. The two specimens collected from S. nigrum were also distinct from those collected on orange trees and fell within the overall variation of the cotton specimens. Although a morphological differentiation based on plant species could be suggested, additional measurements from orange orchards in different locations and cotton populations from Adana Kozan would be required for conclusive results.


12S rRNA analyses
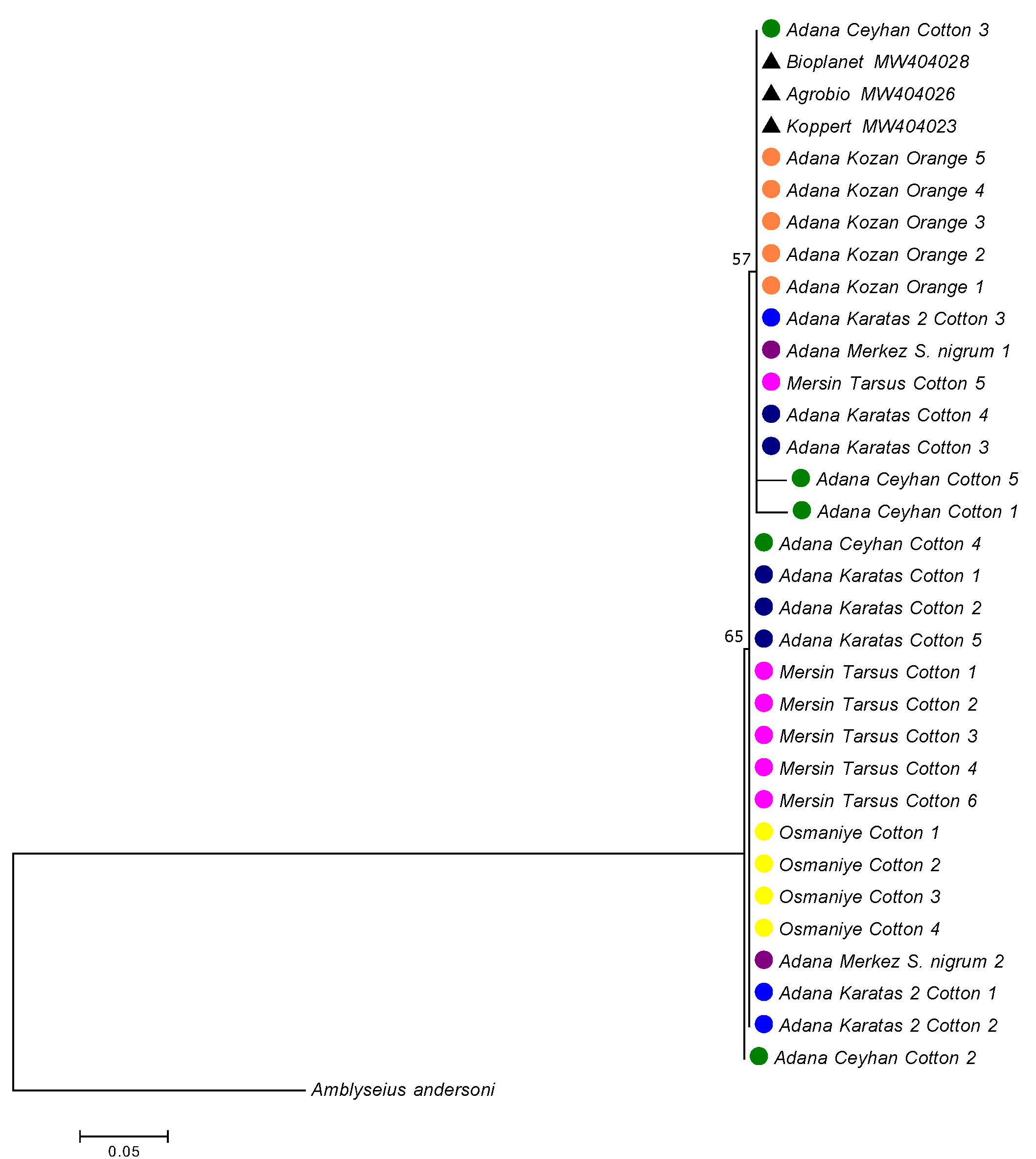


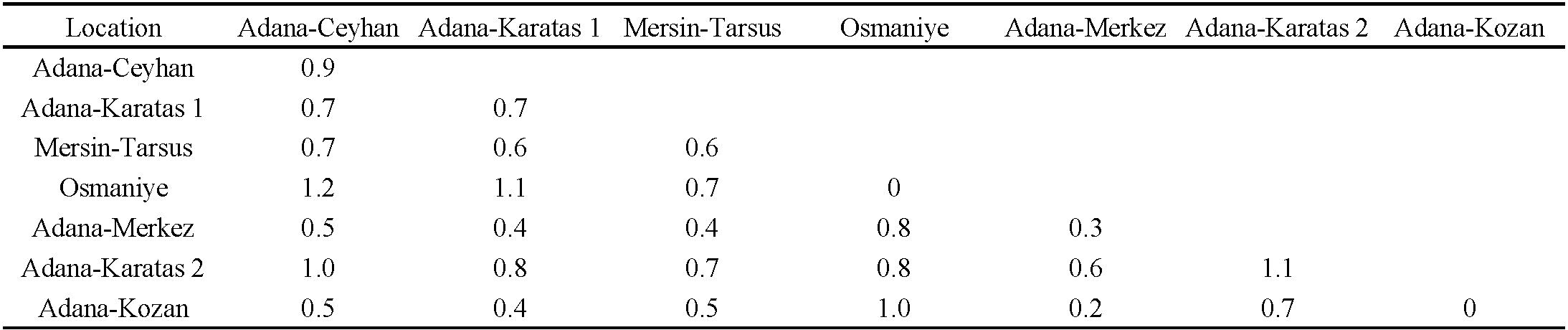


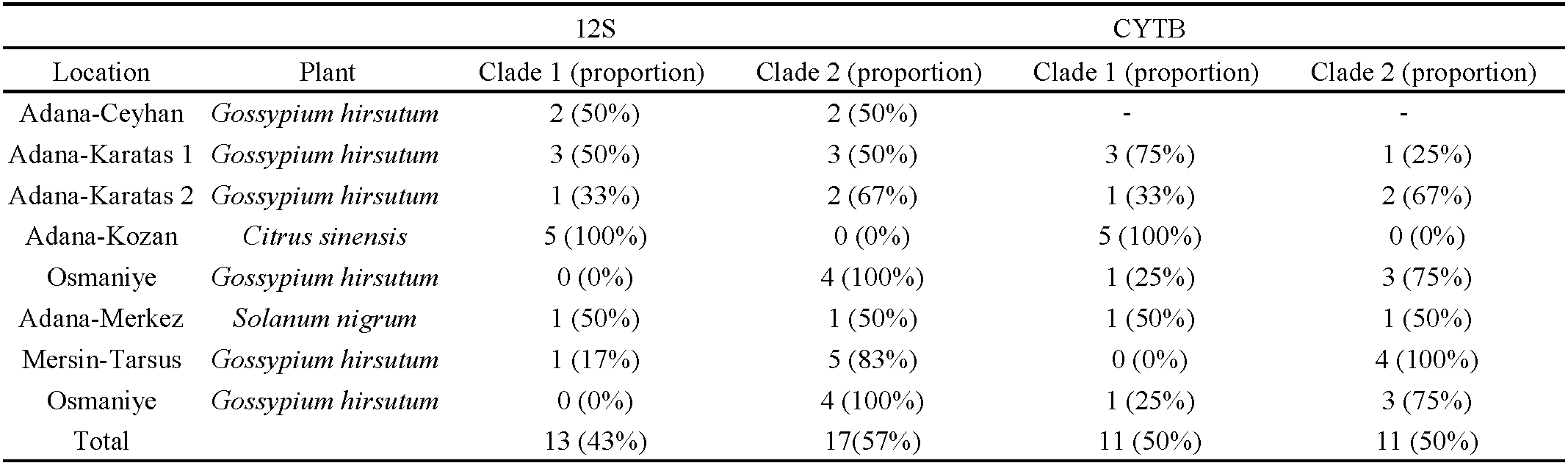
A total of 30 12S rRNA sequences from A. swirskii were obtained. The genetic distances between these 30 specimens were low (0.7% in mean) ranging from 0 to 2.8% (Table 2). The mean genetic distances within populations ranged from 0% (for the Osmaniye population on cotton) to 1.1% (for the Adana-Karatas 2 population on cotton). The genetic distances between populations were quite similar to intra-populations distances, ranging from 0.2% to 1.2%. The lowest genetic distances were observed between the populations collected in Adana Merkez on S. nigrum and in Adana Kozan on orange, while the highest distances were found between the populations from Adana-Ceyhan on cotton and those from Osmaniye and Adana Karatas 2 on cotton. The population from Osmaniye also differed from the populations of Karatas 1 on cotton and Adana Kozan on orange, with a mean genetic distance of 1%. When specimens were grouped according to the three plant species, the mean genetic distance within the ''cotton group'' was 0.8%, within the ''S. nigrum population'' was 0.3%, and within the ''orange group'' was 0%. The mean genetic distance was 0.6% between the ''orange group'' and ''cotton group'', 0.2% between the ''orange group'' and ''S. nigrum population'', and 0.5% between the ''cotton group'' and ''S. nigrum population''. These results suggest that genetic variations were not related to host plants or their geographic locations. The phylogenetic tree revealed two clades, despite the low bootstrap values (Figure 3). The first clade included all specimens collected from orange trees in Adana Kozan, specimens from cotton in Adana Ceyhan (two specimens), Adana-Karatas 1 (three specimens), Mersin-Tarsus (one specimen), Adana Karatas 2 (one specimen), and one specimen from S. nigrum in Adana-Merkez. The second clade contained all specimens collected from cotton in Osmaniye, specimens from cotton in Adana Ceyhan (two specimens), Adana-Karatas 1 (three specimens), Mersin-Tarsus (five specimens), Adana Karatas 2 (two specimens), and one specimen from S. nigrum in Adana-Merkez. The mean genetic distance between these two clades was 0.9%, while the genetic distances within clade 1 and clade 2 were of 0.4% and 0.5%, respectively. Table (3) shows the proportion of specimens per population within these two clades, revealing the following trends: (i) populations with a majority of specimens in clade 1: Adana Kozan on orange, (ii) populations with a majority of specimens in clade 2: Mersin Tarsus on cotton, Osmaniye on cotton and Adana Karatas on cotton, and (iii) populations with specimens equally distributed in the two clades: Adana Ceyhan on cotton, Adana Karatas 1 on cotton, and Adana Merkez on S. nigrum. Specimens from commercial populations sold by Koppert, Agrobio, and Bioplanet were included in the clade 1.


Cytb mtDNA Analyses
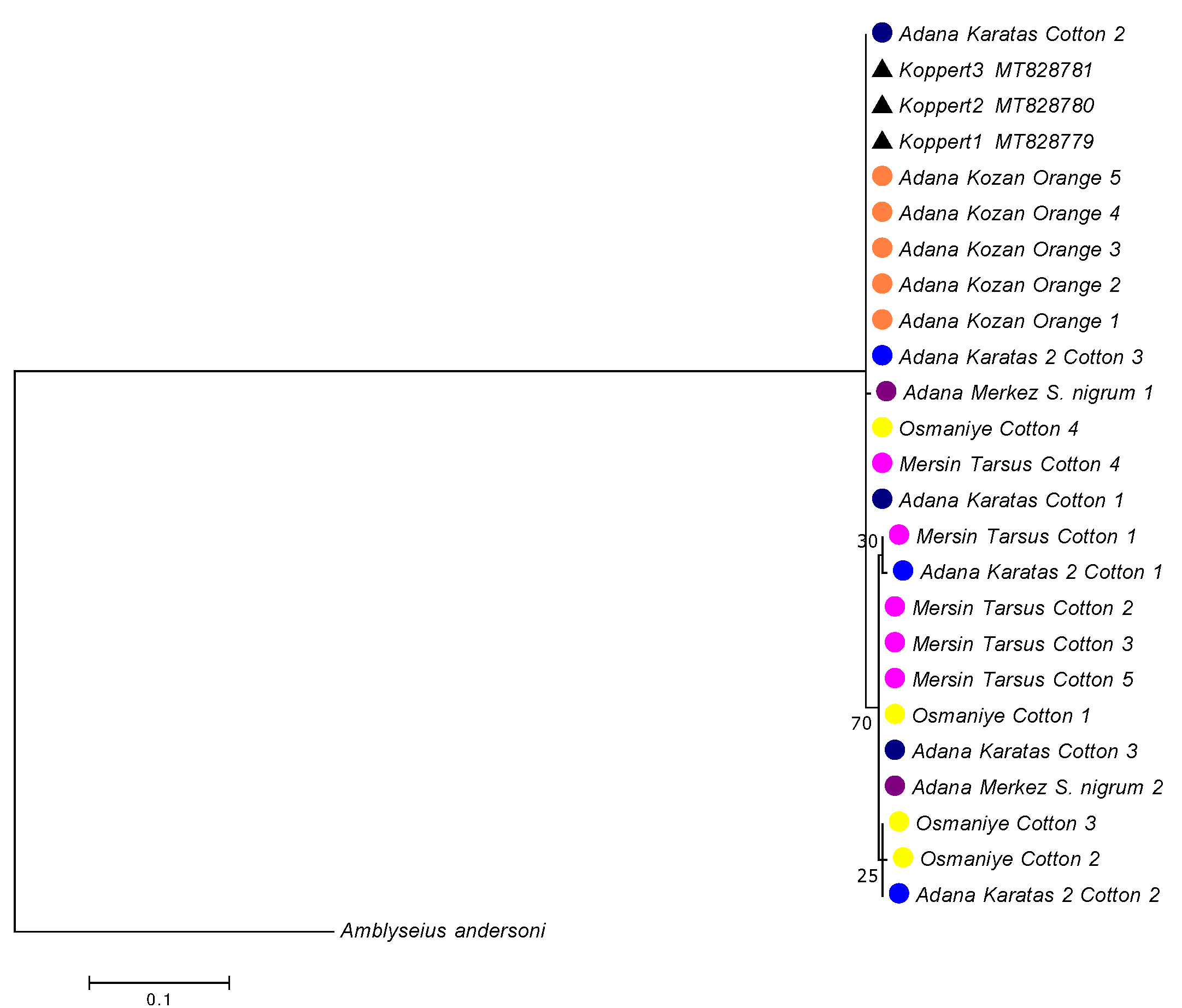


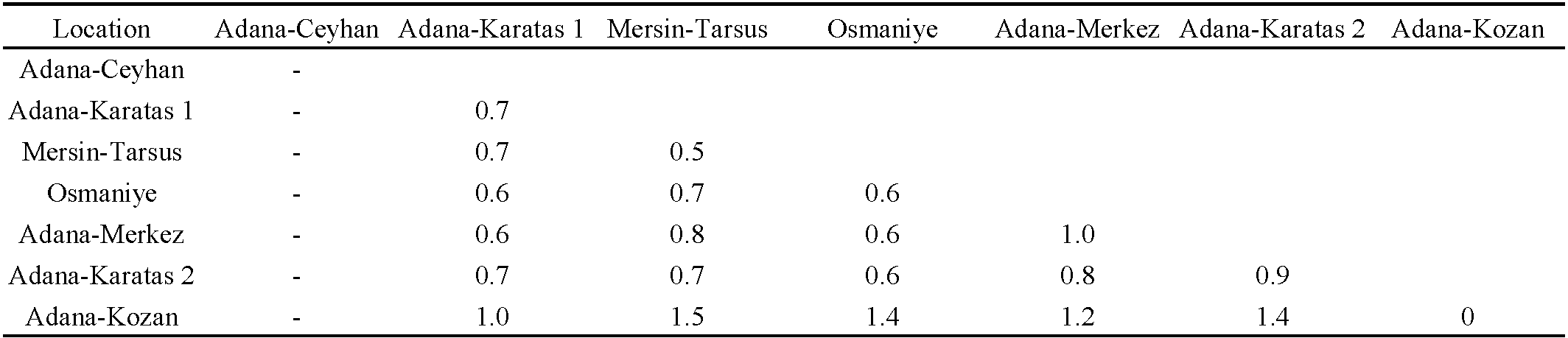


A total of 22 Cytb mtDNA sequences from A. swirskii were obtained. The genetic distances among these speciemens were low, with a mean of 0.9%, ranging from 0 to 1.5% (Table 4). The mean genetic distances within populations ranged from 0% (for the ''Adana Kozan population on orange'') to 1% (for the ''Adana-Merkez'' population on S. nigrum). The mean genetic distances between the populations ranged from 0.6% to 1.5%. The genetic distances between the Adana Kozan population on orange and the other populations were higher than 1%, whereas the genetic distances among the other six populations were lower than 0.8%. When the specimens were grouped according to the three plant species, the mean genetic distance within the ''cotton group'' was 0.7%, within the ''S. nigrum population'' was 1.0%, and within the ''orange group'' was 0%. The genetic distances between these groups were 0.7% between the ''S. nigrum population'' and ''cotton group'', and 1.4% and 1.2% between the ''orange group'' and the ''cotton group'' and ''S. nigrum population'', respectively. As observed with the 12S rRNA fragment, the phylogenetic tree revealed two clades, despite the low bootstrap values (Figure 4). The first clade included all specimens collected on orange trees in Adana Kozan, as well as specimens collected on cotton in Adana-Karatas 1 (three specimens), Osmaniye (one specimen), Adana Karatas 2 (one specimen), and one specimen collected from S. nigrum in Adana-Merkez. The second clade contained specimens collected on cotton in Osmaniye (four specimens), Adana-Karatas 1 (one specimen), Mersin-Tarsus (four specimens), Adana Karatas 2 (two specimens), and one specimen from S. nigrum in Adana-Merkez. The genetic distance between these two clades was 1.3%, while the genetic distances within the clade 1 and clade 2 were 0.8% and 0.4%, respectively. Table (3) shows the proportion of specimens per population within these two clades, revealing the following trends: (i) populations with a majority of specimens in clade 1: Adana Kozan on orange and Adana Karatas 1 on cotton, (ii) populations with a majority of specimens in clade 2: Mersin Tarsus on cotton, Osmaniye on cotton and Adana Karatas 2 on cotton, and (iii) populations with specimens equally distributed in the two clades: Adana Merkez on S. nigrum. These clades contain the same specimens as those included in the two clades obtained with the 12S rRNA fragment, showing concordant results with the two DNA fragments analyzed. Specimens from commercial populations sold by Koppert were included in clade 1, as observed in the phylogenetic tree based on the 12S rRNA fragment. Notably, all specimens collected in orange orchards at Adana Kozan, that were morphologically differentiated from the other populations, were included in the genetic clade 1, indicating a concordance between the morphological and molecular results.
Discussion
Measurements of the A. swirskii females examined are close to those reported by Zannou et al. (2007), Zannou and Hanna (2011), Abo-Shnaf and Moraes (2014), Kreiter et al. (2016), and Döker et al. (2020). Additionally, the min and max values of each character mostly overlapped between all populations. The multifactorial analysis revealed substantial variability within the populations, with the exception of the population from orange at Adana Kozan, which displayed comparatively less variation. It is interesting to note that this population was also genetically differentiated, with all its specimens included in the genetic clade 1, along with commercial specimens. It would be thus interesting in further studies to consider measurements of commercial specimens to assess their similarities with the Adana Kozan population from orange. We can also hypothesize that the morphological differentiation of the population collected on orange trees might be due to a plant effect, as already observed for other phytoseiid species (Schmidt 2014; Tixier 2018; Tixier et al. 2020). Additional studies, including the measurements of specimens from orange orchards and also from other cotton populations from Adana Kozan, would thus be required to draw more definitive conclusions.
The sequences of 12S rRNA and Cytb mtDNA from seven populations of A. swirskii, collected from various locations in southern Türkiye, are reported here for the first time. Despite the identification of approximately 140 phytoseiid species morphologically in Türkiye (Faraji et al. 2011; Döker et al. 2016, 2017a,b, 2018, 2019, 2020; Demite et al. 2024), molecular data are still limited. The genetic distances both for the 12S rRNA and Cytb mtDNA fragments were low for A. swirskii from Türkiye (0.7% for 12S rRNA and 0.9% for Cytb mtDNA). These findings are in line with findings from Tixier et al. (2022b), who reported mean genetic distances of 0% to 1% for 12S rRNA and 0.1% to 1% for Cytb mtDNA among A. swirskii populations from various countries including Egypt, Israel, Reunion Island, Benin and commercial populations. The low genetic distances suggest low genetic variation within A. swirskii, which might indicate high dispersal and gene flow among populations. However, despite these low genetic distances, two distinct clades were observed in the phylogenetic trees using both DNA fragments. There was no clear correlation between these clades and the seven populations of A. swirskii. However, some populations were found exclusively in one of the clades, such as the population from orange trees in Adana Kozan in clade 1 and the population from cotton in Osmaniye in clade 2. However, clades 1 and 2 contained a mixture of specimens from different populations, although varying in proportions. This structuration does not seem to be solely based on locations or the plant species from which the specimens were collected. Other ecological factors might explain this differentiation, such as feeding habits as observed for P. longipes (Tixier et al. 2010a) or climatic conditions, as observed for P. macropilis (Queiroz et al. 2021). Another hypothesis is that differentiation might be associated to differences between natural and commercial specimens. The present results show that the commercial specimens sold by various companies were similar and included in the clade 1, together with the Adana Kozan orange population and some specimens from other populations. Predatory mites, because released in large quantities for biological control purposes, can become more prevalent than naturally occuring species. The observed low differentiation within A. swirskii, as noted by Tixier et al. (2022b), might be attributed to introductions and the ''invasion'' of the commercial specimens. Tixier et al. (2008b) also proposed such an hypothesis to explain the low genetic distances between commercialized specimens and populations of N. californicus collected all over the World. However, this hypothesis would imply a high dispersal abilty of A. swirskii through great distances, as no commercial introduction was reported in this region (I. Döker, pers. comm.).
Conclusion
This study is the first to provide DNA sequences 12S rRNA and Cytb mtDNA from seven A. swirskii populations in southern Türkiye. The analysis revealed low genetic distances between populations, indicating limited genetic variation within this species in Türkiye. Phylogenetic analyses identified two clades with no clear association to geography or host plants. An other hypothesis is that this differentiation might be related to differences between ''commercial specimens'' and ''natural ones''; however no introduction of commercialized population is reported in the regions where the samples were taken. To confirm this hypothesis, it would be interesting to carry out additional studies, taking into account specimens of A. swirskii collected from uncultivated areas and also from other regions of Türkiye where commercial introductions of A. swirskii are reported. The analysis of morphological traits seem to show a differentiation of populations according the host plant; again additional studies with more plants would be required to determine how plant can affect A. swirskii phenotype, taking into account also other characters as the measurements of macrosetae on legs other than leg IV and the ventral setae JV5.
Acknowledgements
The authors acknowledge the support from the Adana Biological Research Institute for their assistance in the collection of the phytoseiids. The authors are deeply grateful for the generous support from the Tübitak 2214 program, a scholarship provided by TÜBİTAK (The Scientific and Technological Research Council of Türkiye). We are also thank the two reviewers for their valuable comments.
References
- Abo-Shnaf R., Moraes G.J. de 2014. Phytoseiid mites (Acari: Phytoseiidae) from Egypt, with new records, descriptions of new species, and a key to species. Zootaxa, 3865(1): 1-71. https://doi.org/10.11646/zootaxa.3865.1.1
- Athias-Henriot C. 1957. Phytoseiidae et Aceosejidae (Acarina, Gamasina) d'Algérie. I. Genres Blattisocius Keegan, Iphiseius Berlese, Amblyseius Berlese, Phytoseius Ribiga, Phytoseiulus Evans. Bull. Soc. Hist. Nat. Afrique du Nord, 48: 319-352.
- Buitenhuis R., Murphy G., Shipp L., Scott-Dupree C. 2015. Amblyseius swirskii in greenhouse production systems: a floricultural perspective. Exp. Appl. Acarol., 65(4): 451-464. https://doi.org/10.1007/s10493-014-9869-9
- Castagnoli M., Falchini L. 1993. Suitability of Polyphagotarsonemus latus (Banks) (Acari Tarsonemidae) as prey for Amblyseius californicus (McGregor) (Acari Phytoseiidae). Redia, 76(2): 273-279.
- Chant D.A., McMurtry J.A. 2007. Illustrated Keys and Diagnoses for the Genera and Subgenera of the Phytoseiidae of the World (Acari: Mesostigmata). IPH, West Bloomfield, 220 pp.
- Demite P.R., Moraes G.J. de, McMurtry J.A., Denmark H.A., Castilho, R.C. 2024. Phytoseiidae Database. [Internet]. [4 July 2024]. Available from: http://www.lea.esalq.usp.br/phytoseiidae
- Döker İ., Kazak C., Karut K. 2016. Contributions to the Phytoseiidae (Acari: Mesostigmata) fauna of Turkey: morphological variations, twelve new records, re-description of some species and a revised key to the Turkish species. Syst. Appl. Acarol., 21(4): 505-527. https://doi.org/10.11158/saa.21.4.10
- Döker İ., Kazak C., Karaca M.M., Karut K. 2017a. Two new records of the genus Kampimodromus Nesbitt (Acari: Phytoseiidae) for Turkey with a revised key to the World species. Acarologia, 57(2): 355-363. DOI: https://doi.org/10.1051/acarologia/20164160
- Döker İ., Kazak C., Karut K. 2017b. Three new species of the family Phytoseiidae (Acari: Mesostigmata). Zootaxa, 4243 (3): 565-576. https://doi.org/10.11646/zootaxa.4243.3.8
- Döker İ., Kazak C., Karaca M.M., Karut K. 2018. Re-discovery and identification of Iphiseius degenerans (Acari: Phytoseiidae) in Turkey, based on morphological and molecular data. Türk. Biyo. Mücadele Derg., 9 (2): 110-123. https://doi.org/10.31019/tbmd.463913
- Döker İ., Kazak C., Karaca M.M., Karut K. 2019. The genus Graminaseius Chant & McMurtry (Acari: Phytoseiidae) in Turkey with descriptions of two new species and re-description of Graminaseius graminis (Chant). Syst. Appl. Acarol., 24(5): 731-741. https://doi.org/10.11158/saa.24.5.2
- Döker İ., Kazak C. 2019. Non-target effects of five acaricides on a native population of Amblyseius swirskii (Acari: Phytoseiidae). Int. J. Acarol., 45(1-2): 69-74. https://doi.org/10.1080/01647954.2018.1542457
- Döker İ., Kazak C., Karut K. 2020. The genus Amblyseius Berlese (Acari: Phytoseiidae) in Turkey with discussion on the identity of Amblyseius meridionalis. Syst. Appl. Acarol., 25(8): 1395-1420. https://doi.org/10.11158/saa.25.8.4
- Faraji F., Çobanoğlu S., Çakmak I. 2011. A checklist and a key for the Phytoseiidae species of Turkey with two new species records (Acari: Mesostigmata). Int. J. Acarol., 37(sup1): 221-243. https://doi.org/10.1080/01647954.2011.558851
- Jeyaprakash A., Hoy M.A. 2002. Mitochondrial 12S rRNA sequences used to design a molecular ladder assay to identify six commercially available phytoseiids (Acari: Phytoseiidae). Biol. Control., 25(2): 136-142. https://doi.org/10.1016/S1049-9644(02)00056-7
- Kanouh M., Tixier M.-S., Okassa M., Kreiter S. 2010a. Phylogenetic and biogeographic analysis of the genus Phytoseiulus (Acari: Phytoseiidae). Zool. Scr., 39(5): 450-461. https://doi.org/10.1111/j.1463-6409.2010.00439.x
- Kanouh M., Tixier M.-S., Guichou S., Cheval B., Kreiter S. 2010b. Two synonymy cases within the genus Neoseiulella (Acari: Phytoseiidae): Is the molecular evidence so evident? Biol. J. Linn. Soc., 101(2): 323-344. https://doi.org/10.1111/j.1095-8312.2010.01516.x
- Kassambara A., Mundt F. 2020. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses. R Package factoextra Version 1.0.7. https://CRAN.R-project.org/package=factoextra
- Kazak C., Döker İ., Karut K. 2016. Abundance of the phytoseiid mites in citrus ecosystems in Adana Province, Turkey. The 8th Symposium of the EURopean Association of ACarologists; July 11-15; Valencia, Spain.
- Knapp M., van Houten Y.M., van Baal E., Groot T. 2018. Use of predatory mites in commercial biocontrol: current status and future prospects. Acarologia, 58(Suppl): 72-82. https://doi.org/10.24349/acarologia/20184275
- Kostiainen T., Hoy M.A. 1996. The Phytoseiidae as biological control agents of pest mites and insects: a Bibliography (1960-1964). University of Florida, Gainesville, 355 pp. https://doi.org/10.1007/BF00051554
- Kreiter S., Vicente V., Tixier M.-S., Fontaine O. 2016. An unexpected occurrence of Amblyseius swirskii (Athias-Henriot) in La Réunion Island (Acari: Phytoseiidae). Acarologia, 56(2): 175-181. https://doi.org/10.1051/acarologia/20162254
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. 2018. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol., 35(6): 1547-1549. https://doi.org/10.1093/molbev/msy096
- Lê S., Josse J., Husson F. 2008. FactoMineR: A Package for Multivariate Analysi. J Stat Software, 25(1): 1-18. https://doi.org/10.18637/jss.v025.i01
- Lee H.S., Gillespie D.R. 2011. Life tables and development of Amblyseius swirskii at different temperatures. Exp. Appl. Acarol., 53(1): 17-27. https://doi.org/10.1007/s10493-010-9385-5
- Lima D.B., Rezende-Puker D., Mendonça R.S., Tixier M.-S., Gondim M.G.C. Jr, Melo J.W.S., Oliveira D.C., Navia D. 2018. Molecular and morphological characterization of the predatory mite Amblyseius largoensis: surprising similarity between Asian and American populations. Exp. Appl. Acarol., 76(3): 287-310. https://doi.org/10.1007/s10493-018-0308-1
- Lindquist E.E., Evans G.O. 1965. Taxonomic Concepts in the Ascidae, with a Modified Setal Nomenclature for the Idiosoma of the Gamasina. Mem. Entomol. Soc. Can., 97(S47): 5-66. https://doi.org/10.4039/entm9747fv
- McMurtry J.A., Croft B.A. 1997. Life-styles of phytoseiid mites and their roles in biological control. Annu. Rev. Entomol., 42: 291-321. https://doi.org/10.1146/annurev.ento.42.1.291
- McMurtry J.A., Moraes G.J. de, Sourassou N.F. 2013. Revision of the life styles of phytoseiid mites and implications for biological control strategies. Syst. Appl. Acarol., 18(4): 297-320. https://doi.org/10.11158/saa.18.4.1
- Okassa M., Kreiter S., Guichou S., Tixier M.-S. 2011. Molecular and morphological boundaries of the predatory mite Neoseiulus californicus (McGregor) (Acari: Phytoseiidae). Biol. J. Linn. Soc., 104(2): 393-406. https://doi.org/10.1111/j.1095-8312.2011.01717.x
- Queiroz M.C.V., Douin M., Sato M.E., Tixier M-S. 2021. Molecular variation of the cytochrome b DNA and protein sequences in Phytoseiulus macropilis and P. persimilis (Acari: Phytoseiidae) reflect population differentiation. Exp. Appl. Acarol., 84: 687-701. https://doi.org/10.1007/s10493-021-00648-w
- R Core Team. 2021. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Internet]. [1 April 2024]. Available from: https://www.R-project.org/
- Rowell H.L., Chant D.A., Hansell R.I.C. 1978. The determination of setal homologies and setal patterns on the dorsal shield in the family Phytoseiidae (Acarina: Mesostigmata). Can. Entomol., 110(8): 859-876. https://doi.org/10.4039/Ent110859-8
- Schmidt R.A. 2014. Leaf structures affect predatory mites (Acari: Phytoseiidae) and biological control: a review. Exp. Appl. Acarol., 62: 1-17. https://doi.org/10.1007/s10493-013-9730-6
- Tixier M.-S., Kreiter S., Cheval B., Auger P. 2003. Morphometric variation between populations of Kampimodromus aberrans (Oudemans) (Acari: Phytoseiidae). Implications for the taxonomy of the genus. Invertebr. Syst., 17(2): 349-358. https://doi.org/10.1071/IS02004
- Tixier M.-S., Kreiter S., Moraes G.J. de 2008a. Biogeographic distribution of the Phytoseiidae (Acari: Mesostigmata). Biol. J. Linn. Soc., 93(4): 845-856. https://doi.org/10.1111/j.1095-8312.2007.00937.x
- Tixier M.-S., Guichou S., Kreiter S. 2008b. Morphological variation in the biological control agent Neoseiulus californicus (McGregor) (Acari : Phytoseiidae): consequences for diagnostic reliability and synonymies. Invertebr. Syst., 22(4): 453-469. https://doi.org/10.1071/IS07052
- Tixier M.-S., Ferrero M., Okassa M., Guichou S., Kreiter S. 2010a. On the specific identity of specimens of Phytoseiulus longipes Evans (Mesostigmata: Phytoseiidae) showing different feeding behaviours: Morphological and Molecular Analyses. Bull. Entomol. Res., 17(1): 1-11. https://doi.org/10.1017/S0007485309990617
- Tixier M.-S., Okassa M., Liguori M.L., Poinso A., Salerno B., Kreiter S. 2010b. Voucher specimens for DNA sequences of Phytoseiid mites (Acari: Mesostigmata). Acarologia, 50(4): 487-494. https://doi.org/10.1051/acarologia/20101984
- Tixier M.-S., Okassa M., Kreiter S. 2012. An integrative morphological and molecular diagnostics for Typhlodromus pyri (Acari: Phytoseiidae). Zool. Scr., 41(1): 68-78. https://doi.org/10.1111/j.1463-6409.2011.00504.x
- Tixier M.-S., Dos Santos Vicente V., Douin M., Duso C., Kreiter S. 2017. Great molecular variation within the species Phytoseius finitimus (Acari: Phytoseiidae): implications for diagnosis decision within the mite family Phytoseiidae. Acarologia, 57(3): 493-515. https://doi.org/10.24349/acarologia/20174168
- Tixier M.-S. 2018. Predatory mites (Acari: Phytoseiidae) in agro-ecosystems and conservation biological control: a review and explorative approach for forecasting plant-predatory mite interactions and mite dispersal. Front. Ecol. Evol., 6: 192. https://doi.org/10.3389/fevo.2018.00192
- Tixier M.-S., Martinez S., Douin M. 2020. Markers of life history traits: Variation in morphology, molecular and amino acid sequences within Typhlodromus (Anthoseius) recki Wainstein (Acari: Mesostigmata: Phytoseiidae). Biol. J. Linn. Soc., 132(1): 53-73. https://doi.org/10.1093/biolinnean/blaa103
- Tixier M.-S., Auger P., Alain M., Fossoud A., Navajas M., Arabuli T. 2021. Integrated taxonomy supports the identification of some species of Phytoseiidae (Acari: Mesostigmata) from Georgia. Acarologia, 61(4): 824-844. https://doi.org/10.24349/m2Rp-WodG
- Tixier M.-S., Tabary L., Douin M. 2022a. Drivers for mutation in amino acid sequences of two mitochondrial proteins (Cytb and COI) in Phytoseiidae mites (Acari: Mesostigmata). Exp. Appl. Acarol., 88: 1-40. https://doi.org/10.1007/s10493-022-00741-8
- Tixier M.-S., Douin M., Lopes I., Migeon A., Fossoud A., Navajas M. 2022b. Genetic diversity of the predatory mite Amblyseius swirskii Athias-Henriot (Acari: Phytoseiidae) with an overview of its distribution and implications for biological control. Biol. Control., 168: 104841. https://doi.org/10.1016/j.biocontrol.2022.104841
- Zannou I.D., Moraes G.J. de, Ueckermann E.A., Oliveira A.R., Yaninek J.S., Hanna R. 2007. Phytoseiid mites of the subtribe Amblyseiina (Acari: Phytoseiidae: Amblyseiini) from sub-Saharan Africa. Zootaxa, 1550(1): 1-47. https://doi.org/10.11646/zootaxa.1550.1.1
- Zannou I.D., Hanna R. 2011. Clarifying the identity of Amblyseius swirskii and Amblyseius rykei (Acari: Phytoseiidae): are they two distinct species or two populations of one species? Exp. Appl. Acarol., 53(4): 339-347. https://doi.org/10.1007/s10493-010-9412-6



2024-08-09
Date accepted:
2024-10-02
Date published:
2024-11-18
Edited by:
Kreiter, Serge

This work is licensed under a Creative Commons Attribution 4.0 International License
2024 Mertoglu Boz, Gamze; Tixier, Marie-Stéphane; Douin, Martial and Kumral, Nabi Alper
Download the citation
RIS with abstract
(Zotero, Endnote, Reference Manager, ProCite, RefWorks, Mendeley)
RIS without abstract
BIB
(Zotero, BibTeX)
TXT
(PubMed, Txt)



