A new species of Litarachna Walter, 1925 (Acari: Hydrachnidia: Pontarachnidae) from Corozal Bay (Belize), described based upon morphology and DNA barcodes
Montes-Ortiz, Lucia1 ; Goldschmidt, Tom2 ; Vásquez-Yeomans, Lourdes3 and Elías-Gutiérrez, Manuel4
1El Colegio de la Frontera Sur, Avenida Centenario km 5.5, Chetumal 77014, Quintana Roo, México.
2Zoologische Staatssammlung München, Münchhausenstraβe 21, D-81247 München, Germany.
3El Colegio de la Frontera Sur, Avenida Centenario km 5.5, Chetumal 77014, Quintana Roo, México.
4El Colegio de la Frontera Sur, Avenida Centenario km 5.5, Chetumal 77014, Quintana Roo, México.
2021 - Volume: 61 Issue: 3 pages: 602-613
https://doi.org/10.24349/r7no-LudgZooBank LSID: BFB2B682-ED8C-4272-954D-78EDD079A444
Original research
Keywords
Abstract
Introduction
The Pontarachnidae Koenike, composed of two genera, Pontarachna Philippi, 1840 and Litarachna Walter, 1925, is the only water mite family with species occurring in marine habitats. Overall, 23 species of the genus Litarachna have been described (Chatterjee et al. 2019) mainly from the intertidal zone of tropical and temperate oceans including the Pacific, Atlantic, and Indian Ocean (Smit 2002; Pešić et al. 2012; Pešić et al. 2019; Chatterjee et al. 2019). Some species occur in estuarine systems as well as in freshwater, e.g. Litarachna brasiliensis Smit, 2007, while others live in shallow coastal marine waters, and some even extend to depths up to 70 m (Pešić et al. 2014), and one is a planktonic species (Litarachna kamui Uchida, 1935). Only four species are recorded from the tropical west Atlantic Ocean, with three occurring in the Caribbean Sea: L. caribica, L. degiustii and L. lopezae (Pešić et al. 2008; Pešić et al. 2012; Chatterjee et al. 2019). In general, the biology and ecology of this group of water mites is mostly unexplored, and little is known about distributional patterns and endemism, particularly in the marine provinces of the American continent (Chatterjee et al. 2019).
Here we describe a new species of Litarachna from Corozal Bay, an estuarine system in Belize, a region within the Yucatan Peninsula biogeographic zone. This species represents the second reported planktonic pontarachnid (Litarachna) mite. It is the first record for Belize, and we present the first DNA barcodes in a description for a member of this family.
Material and methods
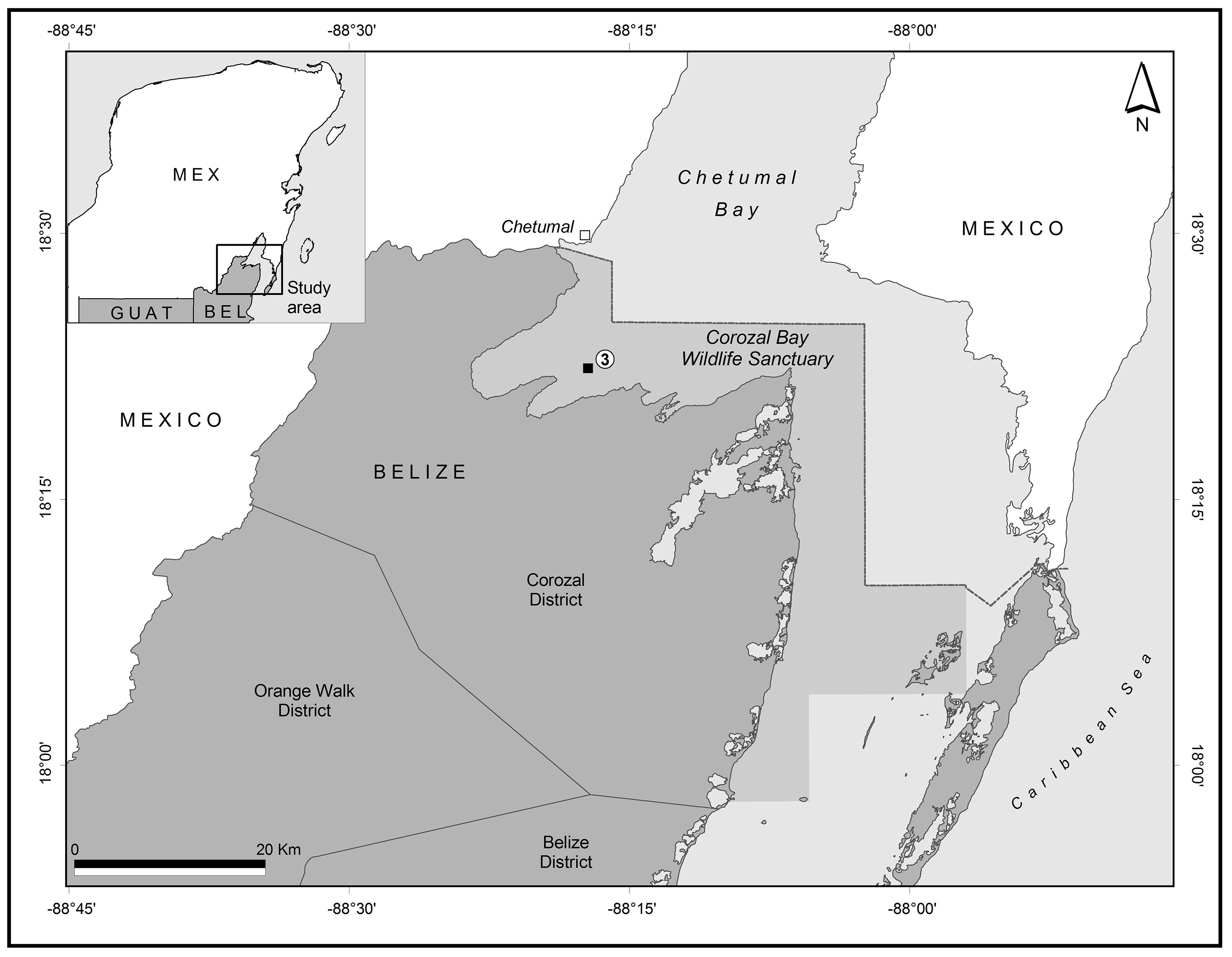


The specimens were collected in open water from Corozal Bay, a binational embayment shared by Mexico and Belize, forming part of Chetumal Bay (Figure 1). A standard plankton net of 0.5 m mouth diameter and a mesh size of 0.333 mm was used to perform a vertical tow during a night sampling survey. The associated data in the collection site are presented in Table 1. Samples were sieved, washed and fixed using 96% ethanol and stored at -18 °C for seven days (Elías-Gutiérrez et al. 2018).
The mites were sorted under a stereomicroscope and one male, and one female were photographed using a Zeiss Discovery stereo microscope with an Eos Rebel T3i camera. SEM images were obtained from a Jeol JSM-SM-6010LA at the Chetumal unit of El Colegio de la Frontera Sur. An additional five individuals were used for molecular analyses using a non-destructive DNA extraction method (Porco et al. 2010). After the process, we recovered three specimens which were dissected and mounted in glycerin jelly. Whole specimens and the dissected parts were examined, measured and drawn under a compound microscope Olympus BX51 with a camera lucida, and the results were compared with prior documentation of this family of water mites (Cook 1996; Smit 2002; Smit 2007; Smit and Alberti 2010; Pešić et al. 2014; Moto and Abé 2014; Chatterjee et al. 2019). All specimens and preparations were deposited in the Reference Collection of Zooplankton at El Colegio de la Frontera Sur (ECOSUR, Chetumal, Mexico).
Molecular analysis. DNA extraction was carried out using a standard glass fiber method (Ivanova et al. 2006) modified following Porco et al. (2010) for voucher recovery. Specimens were recovered after the lysis step from the glass fiber filter plates or the 96 well original plates and preserved in Koenike fluid.
After the DNA extraction, the PCR mixtures contained a final volume of 12.5 μL, including 2 μL of Hyclone ultra-pure water, 6.25 μL of 10% trehalose (previously prepared: 5 g D-(+)-trehalose dehydrate, in 50 ml of total volume of molecular grade ddH2O), 1.25 μL of 10X PCR buffer, 0.625 μL of MgCl2 (50 mM), 0.0625 μL of dNTP (10 mM), 0.125μL of each primer (10μM), 0.06 μL of Platinum Taq DNA polymerase and 2 μL of DNA template. Extracts were amplified with the Zooplankton primers (ZplankF1_t1 and ZplankR1_t1, see Prosser et al. 2013 for details). The reactions were cycled at 94 °C for 1 min, followed by five cycles of 94 °C for 40 sec, 45 °C for 40 sec and 72 °C for 1 min, followed by 35 cycles of 94 °C for 40 sec, 51 °C for 40 sec and 72 °C for 1 min, with a final extension of 72 °C for 5 minutes. PCR products were visualized on a 2% agarose gels (E-Gel 96 Invitrogen), and positive PCR products were selected for sequencing bidirectionally at Eurofins Scientific.
Sequences were edited using Codon Code v. 3.0.1 and uploaded to the Barcode of Life database (BOLD: available at www.boldsystems.org) and are in the public dataset DS-LITBEL. The sequences were included in a maximum likelihood (ML) tree generated with 10,000 replicates using MEGA version X (Kumar et al. 2018); two sequences from L. communis were mined from GeneBank database (https://www.ncbi.nlm.nih.gov/genbank/) and a group of ten sequences of Atractides genus were mined from BOLD to root the tree (Table 2); this genus was selected since they belong to the same Hygrobatoidea superfamily as the pontarachnid mites and include high quality sequences.



All measurements are given in µm. Terminology and abbreviations in the description of the new species follow Moto and Abé (2014) and Gerecke et al. (2016) except for the new abbreviations for the wheel acetabula.
Abbreviations pertaining to morphology of the body: Cx-I-IV = first to fourth coxae, Cxgl-2 = coxoglandularia 2, Cxgl-4 = coxoglandularia 4, L = length, Vgl = ventroglandularia, Vst = ventral setae, W = width, W-1 = wheel acetabula antero-lateral pair, W-2 = wheel acetabula antero-medial pair, W-3 = wheel acetabula posterior pair.
Abbreviations pertaining to appendages (palp, leg): I- to IV-Leg-1-6 = first to sixth segments of leg I to IV, P-1 to P-5 = palp segments 1 to 5.
Abbreviations pertaining to molecular information: BIN = Barcode Index Number, COI = cytochrome oxidase subunit I.
Results
Family Pontarachnidae Koenike, 1910
Genus Litarachna Walter, 1925
Litarachna belicensis n. sp.
ZOOBANK: A09F5614-209C-4446-A6F6-5BE541E56D97 ![]()
(Figures 2–6)













Type series — Holotype: female, Corozal Bay, Belize (18°37´27.7´´ N 88°28´8.44´´ W), 3-5 m depth, substrate mainly calcareous rocks and sand, also sea grass and some spots of fuzzy finger algae (Batophora oerstedi), collection from a planktonic sample from 2.8 m depth, 7 May 2019; dissected and slide mounted in glycerin jelly. Paratypes: one male, same collecting data and processing as holotype.
Diagnosis — Suture between Cx-II and Cx-III incomplete, suture between Cx-III and Cx-IV complete (Figure 6A and D). Postero-medial apodemes twice length of postero-lateral apodemes in both female and male. Ventral projection in P-2 (Figure 3B and 5B), in male very long perigenital setae surrounding the genital field, U-shaped (Figure 5A).
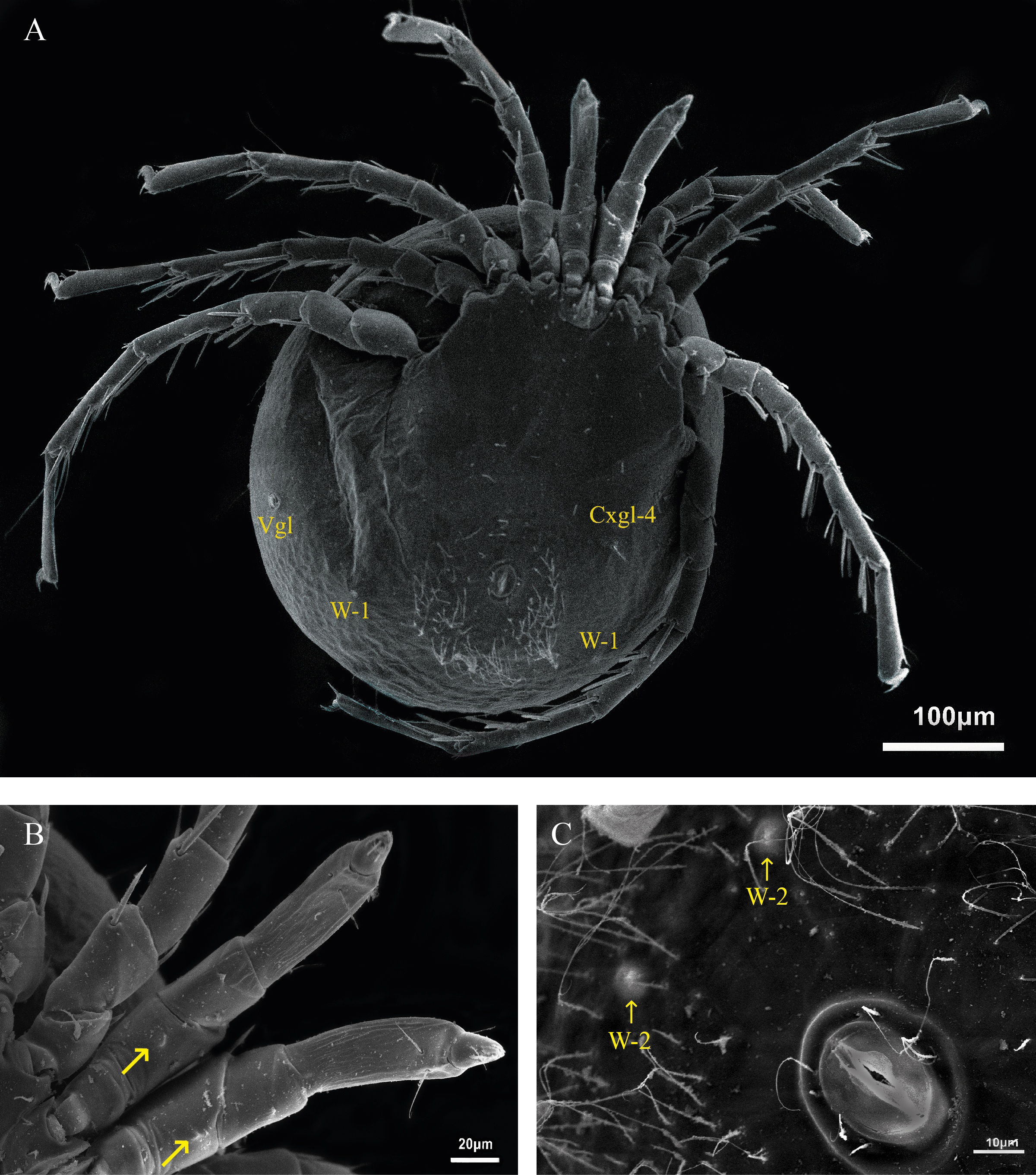
Description — Female (n=1): Idiosoma L/W 476/357. Anterior coxal group separated medially. Suture lines between Cx-II and Cx-III incomplete. Suture lines between Cx-I and Cx-II, and complete between Cx-III and Cx-IV. Posterior margin of Cx-IV with two pairs of long apodemes, extending beyond the genital field, postero-lateral apodemes half as long as postero-medial apodemes (Figures 2, 3, 6A-C)
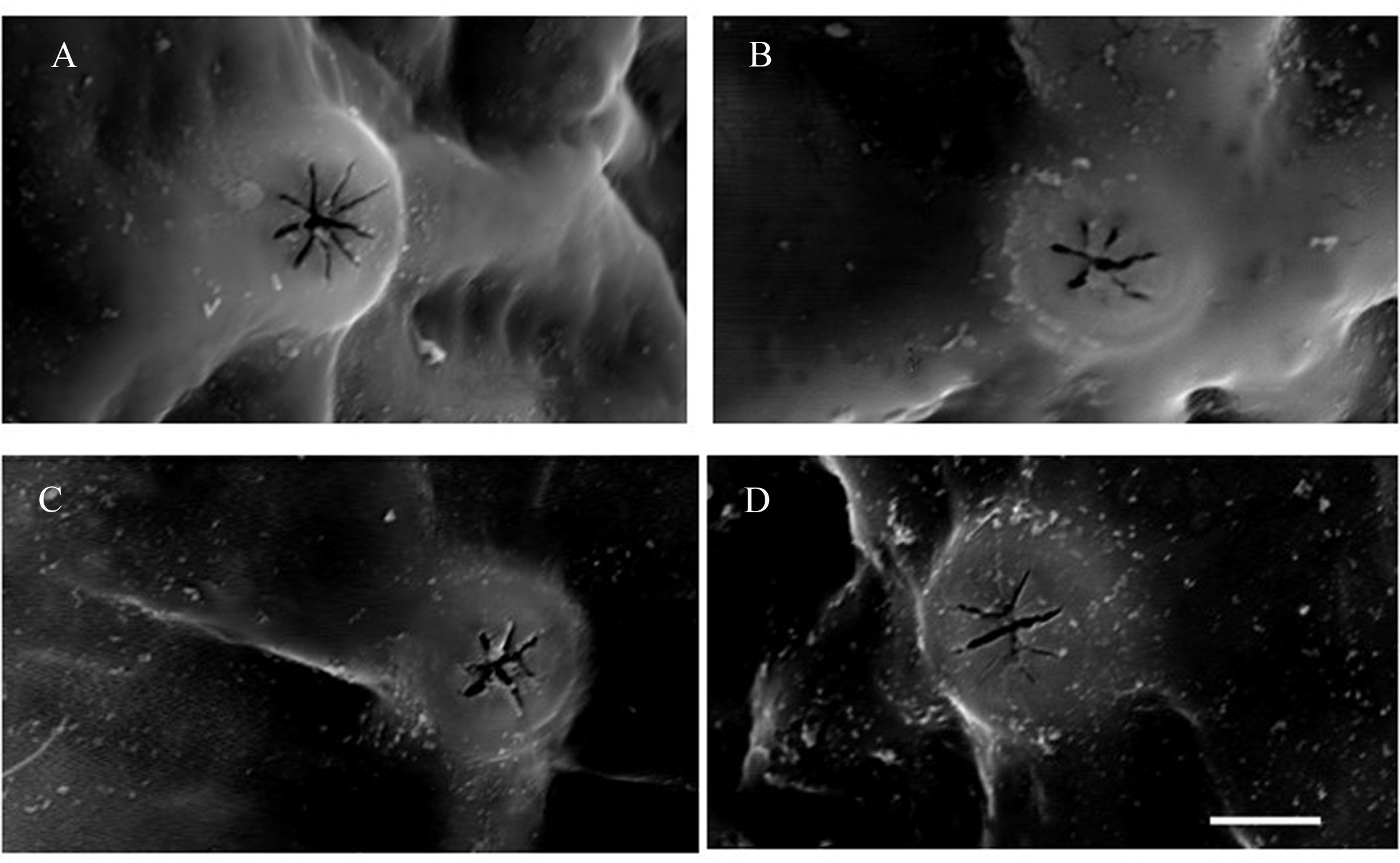
Genital field L/W 78/47. Pregenital and postgenital sclerite fused, forming a ring around genital opening. Cxgl-2 and associated seta (sensu Cook 1974) lie between genital field and the fourth coxae. Posterior to the genital field, a lateral pair of Vgl, and three pairs of wheel-like acetabula (sensu Cook 1996), W-1 with nine radiating spokes, W-2 with eight and seven radiating spokes and W-3 with 10 radiating spokes (Figures 3 and 4). A pair of platelets bearing two pores latero-anteriorly to the excretory pore. Excretory pore sclerotized in subterminal position.
Palp (Figure 3C) total L 194, dorsal L (% of total L): P-1 27 (14%), P-2 33 (17%), P-3 40 (20%), P-4 71 (36%), P-5 25 (13%), P-2 bearing a ventral projection (Figure 3B). L of I-Leg-3-6: 34, 51, 60, 85; II-Leg-3-6: 40, 51, 88, 91; III-Leg-3-6: 28, 52, 95, 85 and IV-Leg-3-6: 57, 95, 119, 104, Leg III and Leg IV each with one swimming seta (Figure 3).
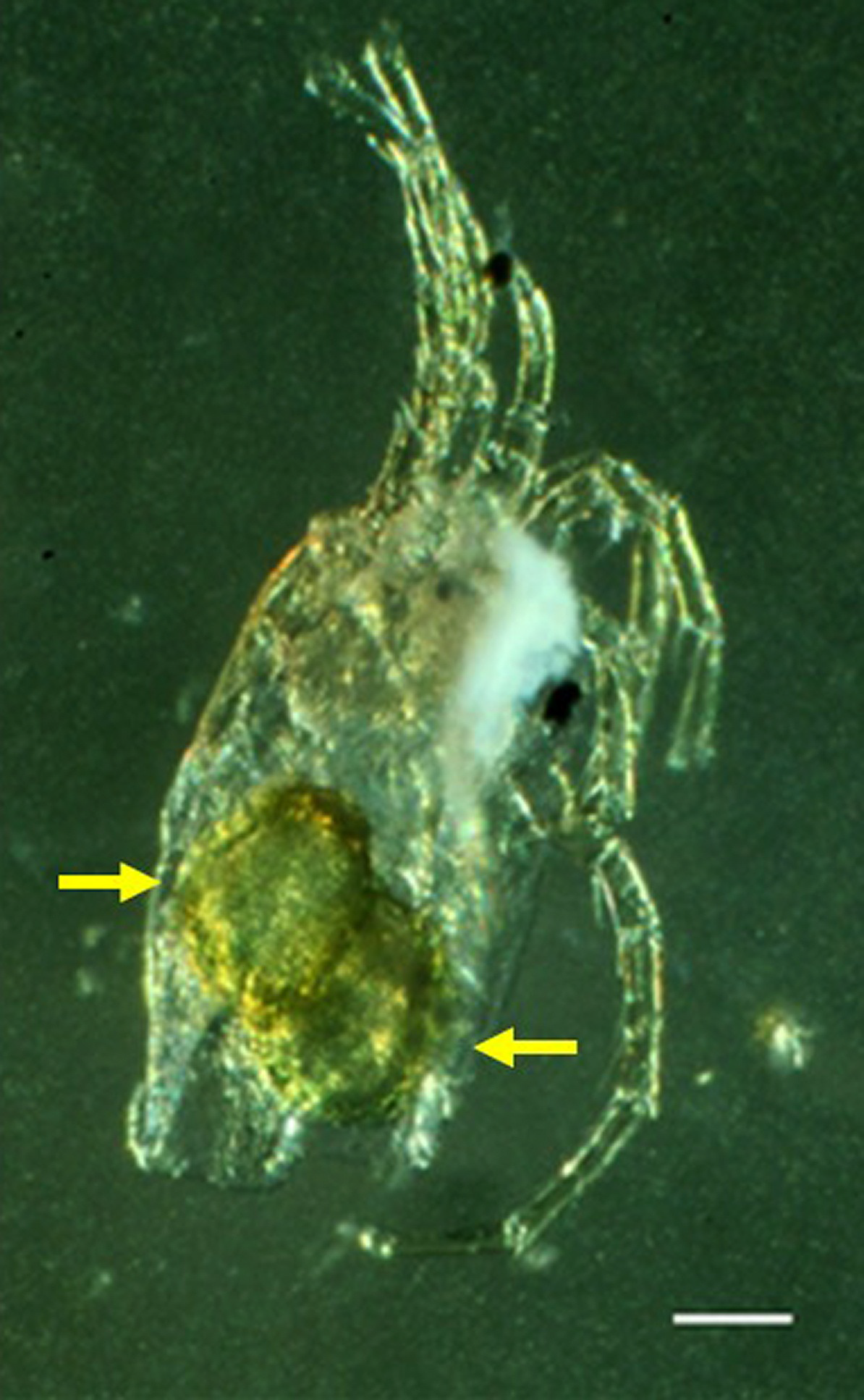
One ovigerous female from the material contained two eggs with a L/W 170/110 (Figure 8).
Male (n=1): Idiosoma L/W 400/348. First coxal plates fused as in the female, suture lines between Cx-II and Cx-III medially incomplete, suture lines complete between Cx-I and Cx-II, as well as between Cx-III and Cx-IV. Postero-medial apodemes twice as long as postero-lateral apodemes, reaching to posterior end of genital field (Figure 6D). Between posterior and lateral apodemes Cxgl-4.
Genital field L/W 35/28, genital sclerites forming a complete ring with four pairs of setae (Figure 5C), many long perigenital setae (\textgreater95) free in the integument surrounding the genital field in a U-shaped form with less dense cover of setae anteriorly and markedly increasing density towards posterior part (Figure 5B); one pair of tiny, wheel-like acetabula at center of perigenital setae, close to the genital opening (Figure 5C). Genital opening flanked by centrally extended lamellae, posterior to the genital field two pores and two pairs of wheel-like acetabula (sensu Cook 1996), W-1, W-3 with nine radiating spokes, W-2 with six and four radiating spokes (Figure 5).
Palp (Figure 5A - B) total L 186, dorsal L (% of total L): P-1 28 (15%), P-2 38 (21%), P-3 40 (22%), P-4 57 (31%), P-5 22 (11%), P-2 bearing a ventral projection (Figure 5B). L of I-Leg-3-6: 39, 50, 64, 54; II-Leg-3-6: 39, 50, 64, 54; III-Leg-3-6: 39, 53, 92, 89 and IV-Leg-3-6: 46, 89, 100, 100.
DNA sequence (CO1) — Two sequences were obtained and resulted in the following consensus sequence:
CTCTATTTTG CTTTAGGAAG ATGATCAGGC ATAATGGGAA CAAGACTTAG 50
AACTTTAATT CGATTAGAAT TAGGTCAACC AGGAGCACTA ATTGGCAATG 100
AACAAATCTA TAACGTTATC GTAACAGCTC ATGCATTTAT TATAATTTTT 150
TTCATAGTCA TACCCATAAT AATTGGAGGT TTTGGAAATT GATTAGTTCC 200
GCTAATAATC AGAGCCCCCG ATATAGCCTT TCCCCGTATA AATAACATAA 250
GATTCTGACT TTTACCCCCA GCCCTTATCC TTCTTTCAAC AAGATCCATA 300
AGATCAATAG GAGCTGGTAC AGGCTGAACA GTTTACCCTC CCCTCTCAAG 350
AAATTTGGCT CACTCAGGAC CATCCGTTGA CTTAACAATC TTCTCTCTCC 400
ATTTAGCTGG TATTTCATCC ATCCTTGGGG CCATCAACTT TATAGCAACA 450
ATTATAAATA TAAAACCTAC CCATATAAAA ATGGAACAAG TACCCCTATT 500
TGTATGATCA ATTTTCATCA CAACCATTCT CCTCCTTCTT TCACTTCCAG 550
TCTTAGCAGG AGCCATTACT ATGCTTTTAA CTGACCGAAA CTTCAACACT 600
TCATTCTTTG ATCCAGCCGG TGGAGGTGAT CCAATTTTAT ACCAACATTT 650
Etymology — This species is named for the country where it was collected.
Discussion
The new species Litarachna belicensis n. sp. is quite similar to L. communis Walter, 1925, originally described in France and with distribution in Croatia, Italy, Montenegro, Turkey (Mediterranean Sea) and Russia (Black Sea), in the complete suture lines between Cx-I and -II as well as Cx-III and -IV and the idiosoma length (L. belicensis 476 – L. communis 465); however, the new species differs from L. communis in the presence of a ventral projection on P-2 and in the absence of two spiniform setae on P-2. Furthermore, the two species differ in the length and width of the apodemes: in L. belicensis postero-medial hook shaped apodemes are wider and twice as long as postero-lateral apodemes, while in L. communis both postero-medial and lateral apodemes are the same length and wide. Litarachna belicensis n. sp. is similar to L. degiustii Cook, 1958 from Bahamas and Netherland Antilles (Caribbean Sea) and L. caribica Pešić et al. 2008 from Panama (Pacific Ocean) and Netherland Antilles (Caribbean Sea), in the shape and size of postero-medial apodemes. The new species differs from these two in the absence of a ventral setal tubercle on P-4, also in the size of postero-lateral apodemes (strongly reduced in the latter two species) and in the state of the suture lines from the coxal plates (both in L. caribica and in L. degiustii suture lines between Cx-II and Cx-III and between Cx-III and Cx-IV are incomplete).



The ML tree generated from COI DNA barcode sequences (Figure 7) reveals that L. belicensis n. sp. is a highly divergent lineage from L. communis, 23% different by K2P. This result agrees with other studies which report similar genetic distances for congeners in the group (Martin et al. 2010; Pešić and Smit 2020). The BOLD grouping algorithm assigns the two species to different Barcode Index Number, BINs: L. communis ADD6045 and L. belicensis n. sp AEB8019 (Ratnasingham and Hebert 2013). These data suggest that L. belicensis n. sp. has a long independent history associated with an ancient separation from the European water mite L. communis, the same could probably be expected for other pontarachnid species in distant marine provinces.


This study is the first to apply an integrative morphological and genetic approach to species delimitation in this family. Our results, in addition to the biogeographic segregation of the new species, confirm the separate identity of L. belicensis as a new species. The integration of molecular data in this study and the future growth of DNA-sequence databases with high quality sequences and complete metadata should contribute to the elucidation of the taxonomic placement of pontarachnid mites, especially the position of Litarachna within the Pontarachnidae as well as of Pontarachnidae within Hydrachnidia. Additional morphological and genetic studies of this enigmatic group will further help to understand their special biology and ecology and to clarify the distribution patterns and degrees of endemism.
Acknowledgements
The financial support to develop this work was provided by the project ''Conservation of Coastal Marine Resources in Central America (Phase II)'', administered by MAR Fund and financed by the Government of Germany through the German Development Bank (KfW). Our gratitude goes to Mr. Joel Verde, Executive Director of Sarteneja Alliance for Conservation and Development (SACD); we also thank the field support from Beatry Verde, Liliany Tamai, Gisel Tepaz, Esmiri Pat, Cesar Muñoz, Honorio Santos, Jose Viamil, and Reynel Blanco.
The result presented here is part of the doctoral investigation research of the first author, being conducted in El Colegio de la Frontera Sur with funds of the National Council of Science and Technology (CONACYT). We thank Alma Estrella García Morales from the Chetumal Node of MEXBOL who assisted with molecular analysis, Holger Weissenberger, from El Colegio de la Frontera Sur, for the elaboration of the map and Harry Smit who provided us fasta files from L. communis. We are in debt with Benjamín C. Víctor and Monica R. Young who kindly performed a style and english review of the manuscript.
We are grateful to anonymous reviewers and Joanna Mąkol for their constructive comments which greatly improved this work.
References
- Chatterjee T., Schizas N. V., Pešić V. 2019. A checklist of Pontarachnidae (Acari: Hydrachnidia) and notes on distributional patterns of the species. Zootaxa, 4619(3): 527-544. https://doi.org/10.11646/zootaxa.4619.3.6
- Cook D. 1996. A freshwater species of Pontarachna, (Acari: Pontarachnidae) from South Africa with a discussion of genital acetabula in the family. An. Inst. Biol. Univ. Nac. Auton. México, Ser. Zool., 67(2): 259-264.
- Elías-Gutiérrez M., Valdez-Moreno M., Topan J., Young M.R., Cohuo-Colli J.A. 2018. Improved protocols to accelerate the assembly of DNA barcode reference libraries for freshwater zooplankton. Ecol. Evol., 8(5): 3002-3018. https://doi.org/10.1002/ece3.3742
- Gerecke R., Gledhill T., Pešić V., Smit H. 2016. Chelicerata: Acari III. In: Gerecke R., ed. Süßwasserfauna von Mitteleuropa, Bd. 7/2-3. Springer-Verlag Berlin, Heidelberg, pp. 1-429. https://doi.org/10.1007/978-3-8274-2689-5
- Ivanova N. V., Dewaard J.R., Hebert P.D.N. 2006. An inexpensive, automation-friendly protocol for recovering high-quality DNA. Mol. Ecol. Notes, 6(4): 998-1002. https://doi.org/10.1111/j.1471-8286.2006.01428.x
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. 2018. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol., 35(6): 1547-1549. https://doi.org/10.1093/molbev/msy096
- Martin P., Dabert M., Dabert J. 2010. Molecular evidence for species separation in the water mite Hygrobates nigromaculatus Lebert, 1879 (Acari: Hydrachnidia): Evolutionary consequences of the loss of larval parasitism. Aquat. Sci., 72(3): 347-360. https://doi.org/10.1007/s00027-010-0135-x
- Moto A., Abé H. 2014. Litarachna communis Walter, 1925 (Acari: Hydrachnidiae: Pontarachnidae): Taxonomic status, lectotype and paralectotype designation and redescription. Acarologia, 54(2): 201-219. https://doi.org/10.1051/acarologia/20142128
- Pešić V., Chatterjee, Tapas, Schizas N. V. 2008. Marine water mites (Acari: Hydrachnidia: Pontarachnidae) from the Caribbean sea, with description of one new species. Cah. Biol. Mar., 49(3): 253-259.
- Pešić V., Chatterjee T., Alfaro M., Schizas N. V. 2014. A new species of Litarachna (Acari: Hydrachnidia: Pontarachnidae) from a Caribbean mesophotic coral ecosystem. Zookeys, 97(425): 89-97. https://doi.org/10.3897/zookeys.425.8110
- Pešić V., Chatterjee T., Schizas N. V. 2012. A new species of Pontarachna (Acari: Hydrachnidia: Pontarachnidae) from a mesophotic coral ecosystem of Vieques Island, Puerto Rico, Caribbean Sea. Zootaxa, 3440: 63-67. https://doi.org/10.11646/zootaxa.3440.1.3
- Pešić V., Durucan F., Zawal A. 2019. Marine mites (Acari: Hydrachnidia) of the Mediterranean Sea: Descriptions of two new species, key for identification and future prospects. Zootaxa, 4585(3): 501-516. https://doi.org/10.11646/zootaxa.4585.3.6
- Pešić V., Smit H. 2020. Mideopsis milankovici sp. nov. a new water mite from Montenegro based on morphological and molecular data (Acariformes: Hydrachnidia: Mideopsidae). Acarologia, 60(3): 566-575.
- Porco D., Rougerie R., Deharveng L., Hebert P. 2010. Coupling non-destructive DNA extraction and voucher retrieval for small soft-bodied arthropods in a high-throughput context: The example of Collembola. Mol. Ecol. Resour., 10(6): 942-945. https://doi.org/10.1111/j.1755-0998.2010.2839.x
- Prosser S., Martínez-Arce A., Elías-Gutiérrez M. 2013. A new set of primers for COI amplification from freshwater microcrustaceans. Mol. Ecol. Resour., 13(6): 1151-1155. https://doi.org/10.1111/1755-0998.12132
- Ratnasingham S., Hebert P.D.N. 2013. A DNA-Based Registry for All Animal Species: The Barcode Index Number (BIN) System. PLoS One, 8(7). https://doi.org/10.1371/journal.pone.0066213
- Smit H. 2002. Two new species of the water mite family Pontarachnidae (Acari: Hydrachnidia), with a discussion of the taxonomic status of Pontarachna hinumaensis Imamura. Zootaxa, 22: 1-8. https://doi.org/10.11646/zootaxa.22.1.1
- Smit H. 2007. Litarachna brasiliensis n. sp., the first member of the water mite family Pontarachnidae (Acari: Hydrachnidia) from South America. Syst. Appl. Acarol., 12(2): 141-146. https://doi.org/10.11158/saa.12.2.8
- Smit H., Alberti G. 2010. The water mite family Pontarachnidae, with new data on its peculiar morphological structures (Acari: Hydrachnidia). Trends Acarol., 71-79. https://doi.org/10.1007/978-90-481-9837-5_11



2021-04-21
Date accepted:
2021-07-11
Date published:
2021-07-21
Edited by:
Mąkol, Joanna

This work is licensed under a Creative Commons Attribution 4.0 International License
2021 Montes-Ortiz, Lucia; Goldschmidt, Tom; Vásquez-Yeomans, Lourdes and Elías-Gutiérrez, Manuel
Download the citation
RIS with abstract
(Zotero, Endnote, Reference Manager, ProCite, RefWorks, Mendeley)
RIS without abstract
BIB
(Zotero, BibTeX)
TXT
(PubMed, Txt)




